2225
Myocardial Segmentation and Phase Unwrapping for Automatic Analysis of DENSE Cardiac MRI using Deep Learning1Biomedical Engineering, University of Virginia, Charlottesville, VA, United States
Synopsis
DENSE myocardial strain imaging is a method wherein tissue displacement is encoded in the image phase. Myocardial segmentation and phase unwrapping are two key steps in quantitative displacement and strain analysis of DENSE images. Prior DENSE analysis methods for segmentation and phase unwrapping were semi-automated techniques, requiring user intervention. In this study, we developed deep learning (DL) methods for fully automated myocardial segmentation and phase unwrapping for short-axis DENSE images. Quantitative and qualitative evaluations show promising results for the proposed DL-based segmentation and phase unwrapping methods, eliminating all manual steps needed for fully automatic DENSE strain analysis.
Introduction
Displacement encoding with stimulated echoes (DENSE) measures myocardial displacement using the signal phase. For good sensitivity to motion, practical displacement encoding frequencies typically cause phase wrapping. To quantify heart motion and compute myocardial strain, myocardial segmentation and phase unwrapping are two key steps. Currently, motion guided segmentation1 and path-following-based phase unwrapping2 are two widely used semi-automatic techniques (integrated in freely available DENSE analysis software3). With these methods, endocardial and epicardial contours need to be defined by the user at one frame, and then motion guided segmentation uses the encoded motion to project a manually defined region of interest through time. This method often needs further revision and manual correction. In addition, phase unwrapping based on path-following typically requires user-defined seed points. Therefore, there is a need to develop a fully automated method that would eliminate the need for user intervention. We investigated the use of deep learning for segmentation and phase unwrapping.Methods
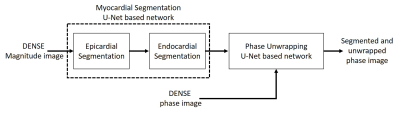
As shown in Fig.1 two networks were trained for myocardial segmentation and another network for phase unwrapping. To segment epicardial and endocardial contours of the left ventricle, a 2D U-Net based structure with dilated convolutions in the contracting path and a combination of weighted cross entropy and soft Dice loss functions was used4. During training, on-the-fly data augmentation including rotation, translation and scaling followed by a b-spline based deformation was used. As reported previously4, to improve the accuracy and smoothness of the segmented contours, during testing, data augmentation by rotating the input images and averaging the probabilistic output was applied. We defined the phase unwrapping problem as a semantic segmentation problem5. As phase wrapping within the myocardium in DENSE is nearly always confined to one cycle, instead of directly obtaining unwrapped phase from wrapped phase, our semantic segmentation network was trained to label each pixel of the myocardial phase image as having no wrap, –2π wrap, or +2π wrap. To create ground truth phase-unwrapped images, we used the path-following method2 to obtain unwrapped phase images, and we manually checked the results, frame by frame, and discarded all frames with unwrapping errors. The same dilated U-Net structure was trained with a pixel-wise cross-entropy loss function. For this network, testing augmentation was not applied, but we applied more training augmentation by adding noise and by manipulating the unwrapped ground truth data to generate new wrapped data. We used DENSE data from 64 subjects for training and from 10 subjects for testing the networks. For the myocardial segmentation network, the training dataset consisted of 6,353 magnitude images, and for the phase unwrapping network the training dataset contained 12,415 short-axis DENSE phase images encoded for motion in the x and y-directions. Network training was performed on an Nvidia Titan Xp GPU over 200 epochs using an Adam optimizer at a learning rate of 5E-4 and a mini batch size of 10. For the test set of 10 subjects, we used the DICE coefficient to compare the similarity between the U-Net and the ground truth. To create the ground truth for myocardial segmentation, our prior semi-automated motion guided segmentation1 followed by an expert user’s manual correction was used.Results
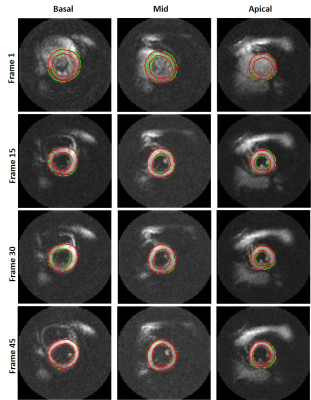
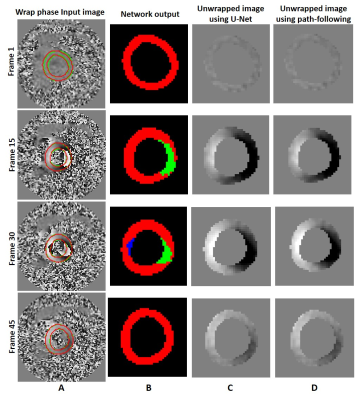
Fig. 2 shows the epicardial and endocardial contours from the ground truth (green) and the network’s output (red) at three slice locations and four temporal frames for one subject. The average dice scores for 1,208 magnitude test images was 88.6% for the segmented myocardium. Moreover, the Dice coefficients for 2,416 test phase images were 99.8%, 95.4%, and 93.8% for pixels without wrap, with –2π wrap, and with +2π wrap, respectively vs. ground truth. Fig. 3 illustrates the phase unwrapping network’s output labels and the computed unwrapped phase images for the U-Net and the path-following method on the segmented myocardium shown in Fig. 2 (mid-ventricular slice).Conclusion
Deep learning accurately performs fully-automated myocardial segmentation and phase unwrapping of cine DENSE short-axis images, eliminating all manual steps needed for fully automatic DENSE strain analysis. Future work will apply these methods to long-axis DENSE images.Acknowledgements
This work was supported by R01HL147104.References
1. Spottiswoode, B.S., et al. "Motion-guided segmentation for cine DENSE MRI." Medical image analysis 13.1 (2009): 105-115.
2. Spottiswoode, B.S., et al. "Tracking myocardial motion from cine DENSE images using spatiotemporal phase unwrapping and temporal fitting." IEEE transactions on medical imaging 26.1 (2007): 15-30.
3. Gilliam, A., DENSEanalysis: Cine DENSE Software. https://github.com/denseanalysis/denseanalysis.
4. Feng, X., et al. “View-independent cardiac MRI segmentation with rotation-based training and testing augmentation using a dilated convolutional neural network”, ISMRM2019.
5. Spoorthi, G. E., et al. "Phasenet: A deep convolutional neural network for two-dimensional phase unwrapping." IEEE Signal Processing Letters 26.1 (2018): 54-58.
Figures