2224
DeepECG: Towards 3-D Continuous Cardiac MRI without ECG-Gating - Deep Learning-based R-Wave Classification for Automated Cardiac Phase Binning1Department of Computer Science, Pattern Recognition Lab, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany, 2Magnetic Resonance, Siemens Healthcare, Erlangen, Germany
Synopsis
For continuous cardiac CINE acquisitions, cardiac binning of the data is necessary, which is done either using ECG-gating or hand-crafted postprocessing methods. To overcome these limitations, we propose a deep learning classifier to detect R-waves from repeated 1-D superior-inferior projections of the imaged data. After training with R-wave positions from the ECG signal as ground-truth data, detection of R-waves is possible without additional ECG-gating or hand-crafted features and can be used for retrospective cardiac binning. Our first proof-of-concept achieves a high accuracy of over 91% on previously unseen cardiac CINE data.
Introduction
Recently, 3-D free-running cardiac scans based on Cartesian or radial spiral sampling were introduced.1-4 Such approaches provide dynamic cardiac volumes, thus, anatomical structures jointly with different cardiac phases1-2 or other information, such as multiple contrasts3 or T1 maps4. To reconstruct this multi-dimensional data, retrospective cardiac and respiratory binning is necessary. By exploiting the repeated k-space center readout of each spiral spoke, which represents a superior-inferior (SI) 1-D projection of the imaged volume, the observation of movements within the volume is possible. Currently, a cardiac phase for every spiral spoke is determined either using ECG-gating2-4 or post-processing such as PCA and ICA1. While ECG-gating requires an additional ECG acquisition, PCA and ICA require prior knowledge, e.g., typical cardiac frequency. To overcome these limitations, we propose a deep learning (DL)-based R-wave detection from SI projections for cardiac binning.Methods
General workflow: We propose the following DL-based workflow (Fig. 1): A temporal window of SI projections is used as input for the DL classifier, which performs a binary classification (“R-wave” vs. “no R-wave”) for every SI projection. Using these predictions, data can be binned into a desired quantity of cardiac phases between two adjacent R-waves. A fully convolutional neural network (FCN) with overall 8 layers is used: Every convolutional layer is followed by a Max-Pooling, reducing spatial dimensionality while preserving temporal resolution. To extract the class labels from the raw output of the network during testing, maximum values from every detected R-wave are taken (Fig. 2).Data acquisition and processing: We acquired data during free-breathing using a 3-D volume-selective, ECG-gated, prototype balanced Steady-State-Free-Precession sequence in short-axis orientation on a 1.5T scanner (MAGNETOM Aera, Siemens Healthcare, Erlangen, Germany). Incoherent subsampling with a spiral spokes pattern was applied to the Cartesian phase-encoding plane.3,5 In order to obtain a broad variability of data, we changed various parameters during our data collection as listed in Tab. 1. The 1-D inverse Fourier-transformed lines of each k-space center readout (SI projections) were used as training input samples, and the simultaneously acquired ECG signal as ground-truth R-wave positions. We divided SI projections into continuous windows of 3 $$$s$$$, each containing multiple and different number of R-wave labels depending on the volunteer’s R-R interval. As all data was different in terms of spatial and temporal resolution as well as number of receiver coils, following preprocessing was applied (resulting in 128x64x10 sized input samples for every 3 $$$s$$$): (1) Temporal interpolation to a fixed size of 64 SI projections for every 3 $$$s$$$ window; (2) Cropping of every SI projection to a fixed spatial resolution (128); (3) Compression of the different coil channels to 10 channels using SVD; and (4) Normalization between 0 and 1 (Fig. 2).
Experiments and Results
Training: We used 17 datasets from 6 healthy male volunteers (25.2$$$\pm$$$9.5 years). To enlarge our training set, data augmentation was conducted by applying randomized cropping sizes from top or bottom of the SI projections and randomized rotations by 180°. We split the data into non-overlapping sets for training (11 datasets, 3 volunteers, 875 windows), validation (2 datasets, 1 volunteer, 73 windows), and testing (2 datasets, 1 volunteer, 257 windows). As overall only few R-waves were present (unbalanced dataset), we used a weighted binary cross entropy loss and optimized with Adam (learning rate: 10-3).6 After training, the model from the epoch with smallest validation loss was used for our tests.Results: Exemplary qualitative results with different heart rates can be seen in Fig. 3. For the quantitative results, we achieve an accuracy of over 91%, on average less than one false positive (FP) and negative (FN) predicted R-wave, and a mean temporal deviation of $$$\approx$$$51 $$$ms$$$ for the correctly predicted R-waves (Tab. 2).
Discussion
Our proof-of-concept delivers a high accuracy of over 91% on previously completely unseen data. Furthermore, the model is capable of handling different heart rates and number of R-waves (Fig. 3 rows 1-3), as well as additional variability introduced by the modified acquisition parameters and the respiratory motion. However, some limitations are existent: First, incorrectly predicted R-waves (FP and FN in Tab. 2) may introduce errors into the binning, leading to inconsistent data for the reconstruction. However, as continuous acquisitions are mostly long 1-4, these single incorrectly binned readouts may cause only minor reconstruction artifacts. Second, the existent mean temporal deviation of correctly predicted R-wave positions in comparison to the ground-truth of $$$\approx$$$51 $$$ms$$$ is within the temporal resolution of one cardiac phase typically used for binning.3 Hence, these readouts will be binned into the same cardiac phase also with this existent deviation. The performance can be increased with more data, especially patient data containing more irregularities compared to volunteers with almost regular heartbeats may be beneficial.Conclusion
We showed a first proof-of-concept for a DL-based detection of R-waves from 1-D SI projections in order to allow an automated, retrospective binning of data into cardiac phases without the need of a simultaneously acquired ECG signal. Providing first promising results, future work will focus on enhancing the performance, including the evaluation on a data base with larger variability or other architectures. Our approach can be easily trained for every existent continuous cardiac sequence using ECG-gating.Acknowledgements
No acknowledgement found.References
[1] Di Sopra, Lorenzo, et al. "An automated approach to fully self‐gated free‐running cardiac and respiratory motion‐resolved 5D whole‐heart MRI." Magnetic Resonance in Medicine 82.6 (2019): 2118-2132.
[2] Kuestner, Thomas, et al.: “3-D Cartesian Free-Running Cardiac and Respiratory Resolved Whole-heart MRI.” Proceedings of the 27th Annual Meeting of ISMRM. Abstract 2192. 2019.
[3] Hoppe, Elisabeth, et al.: “Free-Breathing, Self-Navigated and Dynamic 3-D Multi-Contrast Cardiac CINE Imaging Using Cartesian Sampling and Compressed Sensing.” Proceedings of the 27th Annual Meeting of ISMRM. Abstract 2129. 2019.
[4] Haikun, Qi, et al.: “Free-running 3D Whole Heart Myocardial T1 Mapping with High Isotropic Spatial Resolution.” Proceedings of the 27th Annual Meeting of ISMRM. Abstract 409. 2019.
[5] Wetzl, Jens, et al. "Free-breathing, self-navigated isotropic 3-D CINE imaging of the whole heart using Cartesian sampling." Proceedings of the 24th Annual Meeting of ISMRM. Abstract 411. 2016.
[6] Kingma, Diederik P.,
and Ba, Jimmy: "Adam: A method for stochastic optimization." arXiv
preprint arXiv:1412.6980 (2014).
Figures
Figure 1: DeepECG framework overview
We use every first central spiral spoke k-space readout (1-D inverse Fourier-transformed, marked with orange arrows) as SI projections from the acquisition as small, temporal continuous (3 s) windows for the input of the neural network classifier. The simultaneously acquired ECG signal is discretized to class labels (“R-wave”=1 vs. “no R-wave”=0) and used as ground-truth data for the supervised training.
Figure 2: DeepECG structure
We use SVD-compressed, cropped and temporally interpolated SI projections as inputs. Our FCN consists of 7 blocks with Convolution (3x3 kernel) – ReLU – MaxPooling (3x2 kernel), each increasing the number of feature maps and reducing the spatial resolution by factor 2. The last 1x1 convolution maps the features to one class, followed by a Sigmoid. For the testing, following postprocessing is applied: The output is thresholded at 0.5, the maximum values are taken from every detected R-wave for its exact position and compared with the ground-truth labels.
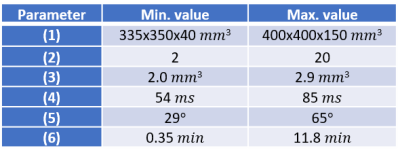
Table 1: Modified acquisition parameters
In order to acquire a broad variability of data, we changed acquisition parameters for every scan during our data collection resulting in the shown ranges of the following parameters: (1) Field-of-View, (2) undersampling factor of the Cartesian phase-encoding plane, (3) spatial resolution, (4) temporal resolution (acquisition time for one spiral spoke), (5) flip angle, (6) total scan time.
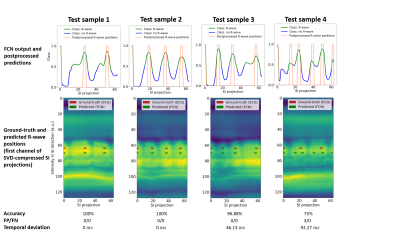
Figure 3: Exemplary qualitative results of one volunteer test dataset
The first 3 columns show that our model predicts R-wave positions on previously unseen data samples and handles different numbers of R-waves and various R-R intervals (columns 1-3). Column 4 shows the worst test case in this volunteer dataset, where additionally false positive R-waves are predicted.
FP: False Positives, FN: False Negatives
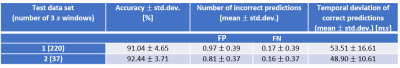
Table 2: Quantitative results for all test datasets
Statistic measures for our test datasets (one long and one short acquisition). The quantitative results show that both scans lead to similar accuracy of over 91% and similar values for incorrectly predicted numbers of R-waves. The mean temporal deviation of correctly predicted R-wave positions of ≈51 ms is still within the typically used resolution of one cardiac phase.
Std.dev.: Standard deviation, FP: False Positives, FN: False Negatives