2207
Towards Realistic Cardiac MR Image Simulation; Inclusion of the Endocardial Trabeculae in the XCAT Heart Anatomy1Biomedical Engineering Department, Eindhoven University of Technology, Eindhoven, Netherlands, 2Carl E. Ravin Advanced Imaging Laboratories, Duke University, Durham, NC, United States, 3Philips Research Laboratories, Hamburg, Germany, 4MR R&D - Clinical Science, Philips Healthcare, Best, Netherlands
Synopsis
The importance of realistic cardiac MR simulation has been realized over the past decade. For this application, XCAT phantom provides realistic highly detailed whole body anatomical models including the heart and respiratory motion. Although the current XCAT heart model is complete in terms of substructures, the trabeculae structure of the endocardium, which is geometrically complex, is lacking. Based on a high-resolution ex-vivo cardiac image data, we modeled and incorporated the irregularity of the trabeculae into the existing XCAT model. We demonstrated that greater realism in cardiac MRI simulation can be achieved by including the trabeculae anatomy into the heart.
Introduction
Magnetic resonance (MR) image simulation plays a crucial role in optimization, benchmarking and validation of medical image analysis algorithms, particularity segmentation methods. Having realistic numerical anatomical models of human organs is one of the essential ingredients of generating realistic simulated MR images. Especially for cardiac application, a highly detailed anatomical model comprising both the heart itself, its substructures, and the surrounding organs with motion functionality needs to be employed. The eXtended CArdiac and Torso (XCAT) phantom1 developed based on Visible Human Project image dataset2 provides a realistic four-dimensional human anatomical model with ability to include cardiac and respiratory motion. The XCAT phantom comprises a high level of anatomical details for the various heart compartments. To improve the phantom realism, the papillary muscles of the left myocardium were included into the XCAT heart phantom. However, the anatomy of myocardial trabeculae for the inner layer of the right and left ventricle of the heart is still lacking in XCAT. The effect of including and excluding these anatomies on the left ventricular quantitative measurements has been assessed3, 4.From the simulation perspective, the importance of the trabeculae inclusion in the appearance of simulated images was previously highlighted by C. Tobon-Gomez et al.5. However, they simulated the presence of the trabeculae by allocating randomly a 3-pixels wide disc to the inner layer of the heart wall and assigning a calculated trabeculae density value rather than modeling the real anatomy. The purpose of this study is to modify the XCAT heart anatomy to increase the realism for simulating cardiac MR images by including realistic trabeculae. To resemble the real anatomy, the trabeculae were precisely modelled based on high spatial resolution cardiac MR image data and integrated into the XCAT. Thereafter, the MR extension to XCAT phantom (MRXCAT6) was utilized to synthesize realistic simulated cardiac MRI. To achieve even more realism, we further extended the MRXCAT to include more than 15 organs around the heart visible in the simulation field of view.
Methods
Modeling of trabeculae: To model the trabeculae of the myocardium, open access ex-vivo high-resolution 3D MRI data of a normal human heart7 were utilized. This whole heart image data was suitable to capture the irregularity of the trabeculae muscular geometry. The trabeculae for right and left ventricle of the heart were manually segmented slice-by-slice using ITK-SNAP software8. A polygon mesh model was exported to be incorporated into the inner surfaces of the XCAT heart chambers. The polygon model for heart’s right ventricle (RV) and left ventricle (LV) was precisely aligned with the heart using Rhinoceros software9.Creation of voxelized phantom: To perform MRI simulation, the creation of a voxelized phantom was performed with XCAT, which includes variable anatomical parameters to generate patient specific anatomical models. The parameters for the torso dimensions in lateral and anterior-posterior directions and the heart orientation within the ribcage were specified based on normal values provided5. The short axis (SA) and 4-chamber (4CH) views of the heart were obtained according to cardiovascular magnetic resonance pocket guide10 by using the rotating and re-slicing functionality of the ImageJ11 processing software tool.
MR simulation: The signal equations based simulation approach of MRXCAT was exploited as the framework of CMR simulation. Cardiac cine MR images were simulated with balanced steady state free precession (b-SSFP) signal model for sequence parameters TR/TE=2.78/1.39, flip angle= 60, 0.5x0.5x4 mm3 voxel size, and 460x460x19 matrix size. To simulate more realistically, we increased the number of structures in MRXCAT by defining additional organ labels available from the voxelized XCAT. MR tissue properties such as T1, T2 and proton density for a normal subject were assigned from literature12.
Results
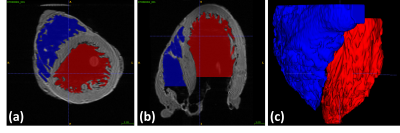
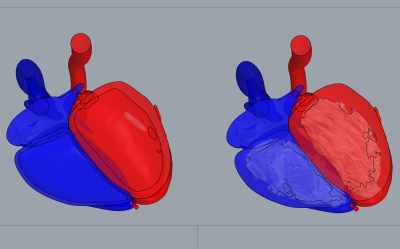
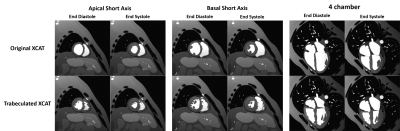
Myocardial trabeculae: Figure 1 shows the manual segmentations of the LV (red) and RV (blue) chambers. A 3D model corresponding to segmented labels is demonstrated such that the tiny jagged-like muscular structure of the trabeculae anatomy is visible. Figure 2 schematically illustrates how the polygon surface mesh model is 3D-aligned with the heart model embedded into the whole XCAT phantom surfaces.MR simulation: The modified version of the XCAT heart clearly improves the anatomical realism of the simulated images. MR simulated images of short axis view and 4-chamber view at end diastolic and end systolic phase of the heart cycle are displayed in Figure 3. To visualize sharply the myocardial trabeculae anatomy, simulation was performed with high spatial resolution.
Discussion
This paper has proposed a step towards more realistic cardiac MRI simulation by modifying the detailed anatomy of the XCAT heart phantom. Particular attention has been paid to model accurately the anatomy of the myocardial trabeculae. To the best of our knowledge, this is the first time that the spatial distribution of the trabeculae has been modeled accurately.Conclusion
We demonstrated that greater realism in cardiac MRI simulation can be achieved by including the trabeculae anatomy into the myocardial wall. In pursuit of generating a database of realistic simulated MR image for medical image analysis research, in our future research we will concentrate on further improving the realism of the MR simulation (including realistic noise, partial volume, tissue texture, etc.).Acknowledgements
This research as a part of the OpenGTN project was supported by the European Union in the Marie Curie Innovative Training Networks (ITN) fellowship program under project No. 764465.References
[1] W. P. Segars, G. Sturgeon, S. Mendonca, J. Grimes, and B. M. W. Tsui, “4D XCAT phantom for multimodality imaging research,” Med. Phys., vol. 37, no. 9, pp. 4902–4915, 2010.
[2] “The Visible Human Project: https://www.nlm.nih.gov/research/visible/visible_human.html.”
[3] J. W. Weinsaft et al., “Left ventricular papillary muscles and trabeculae are significant determinants of cardiac MRI volumetric measurements: Effects on clinical standards in patients with advanced systolic dysfunction,” Int. J. Cardiol., vol. 126, no. 3, pp. 359–365, 2008.
[4] T. Papavassiliu et al., “Effect of endocardial trabeculae on left ventricular measurements and measurement reproducibility at cardiovascular MR imaging,” Radiology, vol. 236, no. 1, pp. 57–64, Jul. 2005.
[5] C. Tobon-Gomez, F. M. Sukno, B. H. Bijnens, M. Huguet, and A. F. Frangi, “Realistic simulation of cardiac magnetic resonance studies modeling anatomical variability, trabeculae, and papillary muscles,” Magn. Reson. Med., vol. 65, no. 1, pp. 280–288, 2011.
[6] L. Wissmann, C. Santelli, W. P. Segars, and S. Kozerke, “MRXCAT: Realistic numerical phantoms for cardiovascular magnetic resonance,” J. Cardiovasc. Magn. Reson., vol. 16, no. 1, p. 63, Dec. 2014.[7] P. A. Helm, H.-J. Tseng, L. Younes, E. R. McVeigh, and R. L. Winslow, “Ex vivo 3D diffusion tensor imaging and quantification of cardiac laminar structure,” Magn. Reson. Med., vol. 54, no. 4, pp. 850–859, Oct. 2005.
[8] P. A. Yushkevich et al., “User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability,” Neuroimage, vol. 31, no. 3, pp. 1116–1128, Jul. 2006.[9] “www.rhino3d.com.”
[10] “Www.cmr-guide.com.”
[11] “www.imagej.nih.gov.”
[12] G. J. Stanisz et al., “T1, T2 relaxation and magnetization transfer in tissue at 3T,” Magn. Reson. Med., vol. 54, no. 3, pp. 507–512, 2005.
Figures