2203
In-vivo cardiac diffusion weighted image registration aided by AI semantic segmentation1Royal Brompton Hospital, London, United Kingdom, 2Imperial College, London, United Kingdom
Synopsis
In-vivo cardiac diffusion weighted images contain contrast differences that complicate image registration from multiple breath-holds. Additionally, neighbouring structures of the chest wall, liver and stomach do not move rigidly with the heart during the respiratory cycle which further hinders registration. In this work we remove other structures and pre-process the image with the help of a convolutional neural network trained to segment multiple heart structures. These additional steps increase the accuracy of registration of the left ventricular myocardial ring resulting in more accurate diffusion tensors.
INTRODUCTION
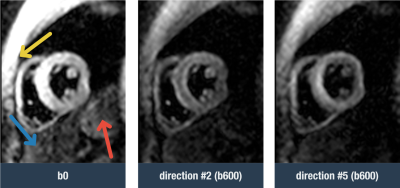
Diffusion tensor cardiac magnetic resonance (DT-CMR) is an emerging technique that provides information on myocardial microstructure. Many different diffusion parameters can be extracted from the tensors, including tensor orientation measures which have been shown to relate to the orientation of the local cardiomyocytes and their sheetlet structure1. In-vivo diffusion imaging yields low SNR images, and it is common to compensate with long acquisitions spanning multiple breath-holds. For most scans an image registration algorithm is needed to correct for residual image shifts due to problems with intra and inter-breath hold stability and consistency. This registration is often challenging due to the different diffusion weightings between images and or different diffusion encoding directions which translate to differences in signal intensities and contrast, not only of the heart muscle, but also the neighbouring structures of the chest wall, liver and stomach (figure 1). Additionally, these peripheral structures do not move rigidly with the heart during the respiratory cycle.Automatic left ventricular myocardial segmentation of DT-CMR images with a trained convolutional neural network (CNN)2 has been recently developed. In this work we hypothesize that we can improve the accuracy of image registration by segmenting the heart and removing other neighbouring structures from the images prior to registration.
METHODS
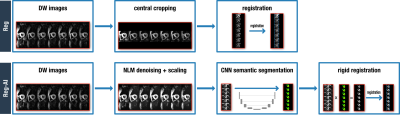
A previously trained U-Net2,3 was modified to include multiple classes, including right ventricular myocardium and papillary muscle, and interventricular insertion points. This modified multi-class U-Net was trained on 393 DT-CMR scans and cross-validated on additional 29 scans, and finally tested on 70 independent scans. All DT-CMR scans were acquired with a STEAM-EPI sequence that were previously segmented by an experienced user.The goal is to use this U-Net to segment the heart in each individual diffusion image prior to registration. In order for this to work reliably, the diffusion images were denoised with a non-local means algorithm and magnitude scaled before applying the U-Net. The resulting segmentation mask was then used to remove all peripheral structures outside the heart and to increase the image intensity towards the LV myocardial region. A rigid image registration was subsequently performed4. This method, referred to here as Reg-AI was compared to the currently used image registration algorithm where only a square crop is done to extract the central part of the field-of-view prior to using the rigid registration algorithm (Reg). A flowchart of these methods is shown in figure 2. The image with the highest mean intensity was used as the reference frame for registration.
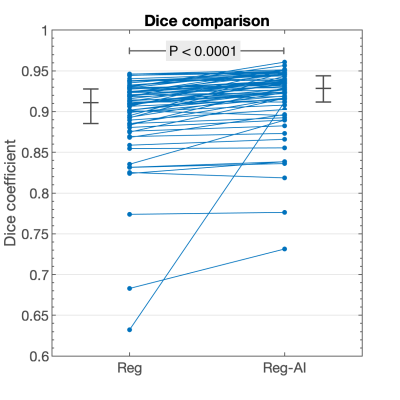
Differences in the two image registration algorithms (Reg vs Reg-AI) were quantified using the registered myocardial masks and calculating the mean Dice coefficient of all the diffusion images in the scan in comparison to the reference frame. A non-parametric Wilcoxon signed rank test was used to quantify statistical difference between the two methods.
RESULTS
The multi-class U-Net was capable of successfully segmenting the individual denoised diffusion frames. A Dice score of 0.93 [0.02] (median [IQR]) was achieved for the LV myocardial ring after registration when compared to the segmentation of the same region in the registered scan average image (higher SNR than the reference frame).There was a significant improvement in the image registration of the new Reg-AI method compared to the current Reg method (P < 0.0001) (figure 3). The median Dice coefficient for Reg and Reg-AI were 0.91 [0.04] and 0.93 [0.03] respectively, this time in relation to the reference frame.
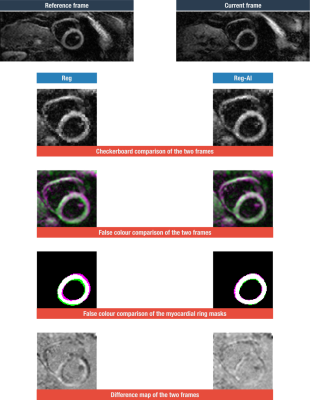
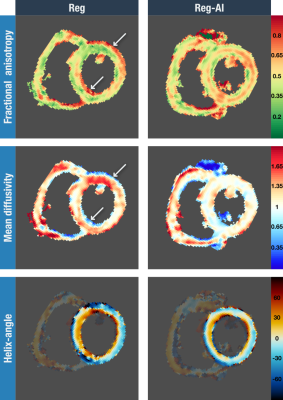
An example where Reg-AI performed noticeably better than the current Reg method is shown in figure 4. The resulting DTI parameter maps, for this particular scan with the two registration methods, are shown in figure 5. Conspicuous artefacts can be seen for the Reg method due to the poorer registration.
DISCUSSION
Using a trained CNN capable of segmenting the heart, and in particular the LV myocardial ring, prior to performing a rigid image registration algorithm improves the registration of in-vivo DT-CMR scans. Improvements are greatest when there are bright structures adjacent to the heart, commonly the chest wall, the liver and the stomach. These organs do not move rigidly with the heart during the respiratory cycle and therefore introduce errors when registering the heart in a series of diffusion weighted images acquired from multiple breath-holds. Refining image registration is important to robustly calculate pixelwise diffusion tensors.Acknowledgements
This work was supported by the British Heart Foundation RG/19/1/34160.References
[1] Nielles-Vallespin S, Khalique Z, Ferreira PF, de Silva R, Scott AD, Kilner P, McGill L, Giannakidis A, Gatehouse PD, Ennis D, Aliotta E, Al-Khalil M, Kellman P, Mazilu D, Balaban RS, Firmin DN, Arai AE, Pennell DJ. Assessment of Myocardial Microstructural Dynamics by In Vivo Diffusion Tensor Cardiac Magnetic Resonance. J Am Coll Cardiol. 2017; 69: 661-676.
[2] Ferreira PF, Scott AD, Khalique Z, Yang G, Nielles-Vallespin S, Pennell DJ, et al. Fully automated DT-CMR analysis with deep learning. In: Proc Intl Soc Magn Reson Med; 2019.
[3] Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF, ed. Medical Image Computing and Computer-Assisted Intervention -- MICCAI 2015; 2015:234–241.
[4] Guizar-Sicairos M, Thurman ST, Fienup JR. Efficient subpixel image registration algorithms. Opt Lett. 2008; 33: 156-8.
Figures