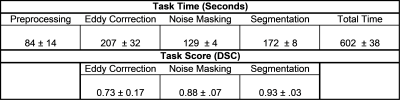2198
SADDLE: A Stand Alone Device for Deep Learning Execution1Northwestern Radiology, Chicago, IL, United States, 2Ann & Robert H. Lurie Children's Hospital of Chicago, Chicago, IL, United States
Synopsis
The integration and evaluation of a stand-alone solution for deep learning execution within clinical environments was tested. Using a small GPU device preloaded with a data listener and pre-trained neural networks, bicuspid aortic valve patients having 4D-flow scans were pre-processed and had their aorta’s segmented automatically. Eddy current correction, noise masking, and aortic segmentation were processed sequentially. The device was integrated within the hospital’s network so that data always stayed on-site. The average time for 4D-flow processing and segmentation was roughly 10 minutes. Dice’s coefficients were 0.72±0.17, 0.87±.07, and 0.93±0.03 for eddy current correction, noise masking, and aortic segmentation.
Introduction
Deep learning convolutional neural networks (CNNs) have shown potential for classification, segmentation, and data processing tasks in medical imaging.1,2 4D-flow MRI provides spatial and time resolved 3D hemodynamic information and has shown to be a promising technique for evaluation of altered cardiovascular hemodynamics in aortic disease such as aortic valve abnormalities, dissection, and coarctation.3–5 However, 4D-flow MRI data requires time intensive, manual post-processing and segmentation of the vessel of interest (e.g. thoracic aorta). In addition, data analysis requires phase offset correction of eddy currents, de-noising, and velocity anti-aliasing prior to 3D segmentation. Recently, we have shown that dedicated deep learning CNNs for each task allows for efficient automated processing with excellent performance compared to the ground truth (manual analysis).6,7 However, these networks tend to extend poorly to other centers and require sufficient hardware and technical expertise, including data management (data retrieval and post-processing storage), data processing (execution of CNN), and efficient integration into a clinical data archiving resources (e.g. VNA,PACs). This stymies inter-site collaboration and testing of models, as data may not be rapidly shared, and limits the broad applicability of deploying deep learning models into a clinical environment. Cloud-based CNN strategies8 have been proposed to offload data processing to suitable CPU/GPU architectures, but these are limited by upload speeds, connection to highly varying local data achieving solutions (e.g. VNA,PACs), and concerns regarding data privacy (data leaving the hospital firewall). To address these limitations, we have developed a stand-alone solution, SADDLE, integrating low-cost hardware with pre-loaded deep learning models and listener system, operating fully inside the hospital network environment and firewall. The purpose of this study was to evaluate its utility for easy integration into any hospital data network for efficient automated 4D-flow analysis workflows.Methods
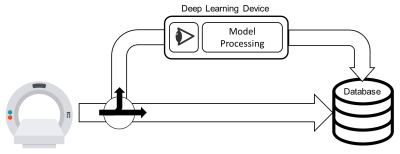
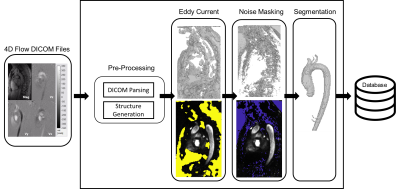
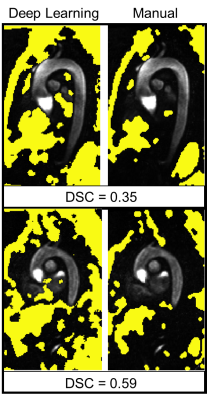
A small GPU device (Jetson Nano, Nvida), 4 GB memory, was integrated into our data retrieval pipeline. The device was pre-loaded with pre-processing deep learning networks (3D noise masking and eddy current correction), aortic segmentation deep learning network, and a customized polling observer (Watchdog, Apache Foundation). The observer was developed to watch incoming patient scan data, process data meeting search criteria, and return processed data back to the processing pipeline (Figure 1). 10 retrospectively selected bicuspid aortic valve patients (48.4±18, 7 Male) were streamed into our observed location. Patients were selected such that they were not utilized for prior CNN training (i.e. true test cases). Scan parameters for 4D-Flow acquisition were: 1.5 Tesla (Avanto, Siemens), Gadavist contrast, GRAPPA R=2, temporal resolution= 36-38.4ms, spatial resolution= 2.125-2.5x 2.125-2.5x 2.6-3.0mm3, venc= 150-250cm/s, and FOV= [277-341mm, 340-400mm, 68-91mm]. 4D-flow aortic scans, composed of DICOM images, were, in real-time, identified and processed. Task times were logged after each completion. Each deep learning task (serial execution of CNNs for noise masking, eddy current correction and 3D segmentation, see Fig 2) was compared to its ground truth analog: a human observer’s processing. For eddy current correction and noise masking, the human observer used an in house processing tool to generate a correction mask. For 3D aortic segmentation, a human observer manually segmented the aorta region of interest (Mimics, Materialise). Dice’s coefficients9 (DSC) were used to quantify the differences between the ground truth and deep learning masks and segmentations (Figure 2).Results
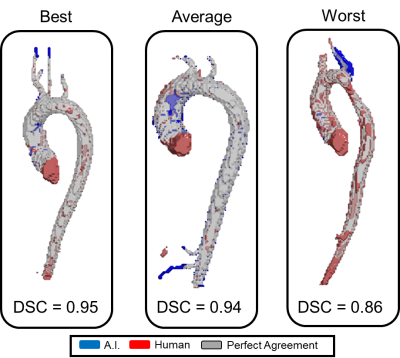
The device was successfully integrated within the hospital network through physical connection and drive mapping. Average total time for fully automated patient processing and result saving was 602±39 seconds. Preprocessing of DICOM images for model input, eddy current correction, noise masking, and segmentation took, on average, 84±14, 207±32, 128±4, and 172±8 seconds, respectively. DSC scores of 0.72±0.17, 0.87±.07, and 0.93±0.03 were achieved between deep learning and manual masks and segmentations for eddy current correction, noise masking, and segmentation respectively (Figures 3,4).Discussion
We were successful in transferring our deep learning models onto a stand-alone GPU device. Importantly, the device integrates within the hospital network and firewall so the data never leaves the on-site network. The device took roughly 10 minutes to automatically process each patient which we believe is clinically feasible; however, velocity anti-aliasing network wasn’t implemented due to memory constraints but will be in future work. Overall, the deep learning models performed well and similar to our prior work except for eddy current correction. This model was trained and validated in a pediatric population and underperformed in our adult patients (which may be mostly due to the coarse tool and non-optimal static tissue choice in the manual processing, see Figure 5). This study underscores the importance and potential of this solution and plug-and-process framework for model evaluation across sites and cohorts. Furthermore, while this study looked specifically at 4D-flow processing and aortic segmentation, the framework can be extended to other imaging modalities, deep learning models and tasks, and quantifications such as deriving and reporting flow or anatomical metrics.Conclusion
We developed an on-site, internal stand-alone system for the utilization of deep learning models to process imaging data. In this study, we examined our preprocessing and aortic segmentation networks for bicuspid aortic valve patients which took, on average, 10 minutes to complete per patient and required no user intervention to complete.Acknowledgements
This research was supported by NIH grant F30HL145995.References
1. Suzuki K. Overview of deep learning in medical imaging. Radiol Phys Technol. 2017;10(3):257-273. doi:10.1007/s12194-017-0406-5
2. Lee JG, Jun S, Cho YW, et al. Deep learning in medical imaging: General overview. Korean J Radiol. 2017;18(4):570-584. doi:10.3348/kjr.2017.18.4.570
3. Garcia J, Barker AJ, Markl M. The Role of Imaging of Flow Patterns by 4D Flow MRI in Aortic Stenosis. JACC Cardiovasc Imaging. 2019;12(2):252-266. doi:10.1016/j.jcmg.2018.10.034
4. François CJ, Markl M, Schiebler ML, et al. Four-dimensional, flow-sensitive magnetic resonance imaging of blood flow patterns in thoracic aortic dissections. J Thorac Cardiovasc Surg. 2013;145(5):1359-1366. doi:10.1016/j.jtcvs.2012.07.019
5. Hope MD, Meadows AK, Hope TA, et al. Clinical evaluation of aortic coarctation with 4D flow MR imaging. J Magn Reson Imaging. 2010;31(3):711-718. doi:10.1002/jmri.22083
6. Berhane H, Scott M, Robinson J, Rigsby C, Markl M. 3D U-Net for Automated Segmentation of the Thoracic Aorta in 4D-Flow derived 3D PC-MRA. In: ISMRM2019. ; 2019. 7. Berhane H, Scott M, Elbaz M, Markl M. Artificial intelligence-based fullly automated 3D segmentation of the aorta from 4D flow MRI. In: SMRA 2019. ; 2019.
8. Yates EJ, Yates LC, Harvey H. Machine learning “red dot”: open-source, cloud, deep convolutional neural networks in chest radiograph binary normality classification. Clin Radiol. 2018;73(9):827-831. doi:10.1016/j.crad.2018.05.015
9. Zou KH, Warfield SK, Bharatha A, et al. Statistical validation of image segmentation quality based on a spatial overlap index. Acad Radiol. 2004;11(2):178-189. doi:10.1016/S1076-6332(03)00671-8
Figures