2177
Myocardial strain of hypertrophic ventricular noncompaction, isolated ventricular noncompaction and hypertrophic cardiomyopathy1Radiology, Union Hospital, Tongji Medical College, Huazhong University of Scince and Technology, Wuhan, China, 2MR Collaboration, Siemens Healthineers Ltd., Shanghai, China
Synopsis
Understanding of ventricular noncompaction appearance with simultaneous hypertrophic cardiomyopathy remains to be improved, as conventional used markers may not be able to distinguish subtle variation between isolated ventricular noncompaction and hypertrophic cardiomyopathy. We utilize the myocardial strain analysis based on cine images to compare different subgroups in order to find clues of underlying pathophysiology. Remarkable difference of the strain value were found in hypertrophic cardiomyopathy patients compared with the isolated ventricular noncompaction ones.
Introduction
Ventricular noncompaction (VNC), whether it is a distinct cardiomyopathy or a morphologic trait, is still on debate[1]. Its association with other cardiomyopathy may present quiet different outcomes[2] from isolated ventricular noncompaction (IVNC). Hypertrophic cardiomyopathy (HCM), the most common genetic cardiomyopathy, may coexist with ventricular noncompaction and may show some distinct features different from the either ones[3]. Cardiac MR derived parameters, such as myocardial strain, are sensitive to detect subtle differences relating to structure configuration[4]. Hypertrophic subtype of ventricular noncompaction has not been evaluated and compared with IVNC and HCM using cardiac MR strain method. The aim of this study was to compare cardiac strain parameters between hypertrophic VNC, IVNC, HCM and normal controls.Methods
We retrospectively enrolled 12 patients with hypertrophic subtype of VNC(age: 43.7±19.1, female 4/10), 15 IVNC patients(age:40.4±16.9, female 5/15) and 20 HCM patients(age:43.0±17.2, female 8/20) in this study. Besides, 20 sex and age matched controls were also collected. Cardiac MR cine imaging was performed using a steady-state free precession sequence on a 1.5T MAGNETOM Area MR scanner (Siemens Healthcare, Erlangen, Germany). Ejection fraction (EF) and strain parameters of left ventricle derived from longitudinal and short axial cine images were measured with CVI 42 software (Circle Cardiovascular Imaging Inc, Calgary, Alberta,Canada) and were compared between groups. Statistical significance was defined as P <0.05.Results
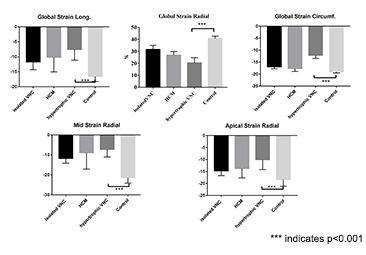
The mean EF showed no significant differences between groups even compared with normal controls. However, compared with IVNC or HCM group, strain parameters between hypertrophic VNC group and control group showed more remarkable difference no matter in global (global longitudinal strain:(-7.57±3.59)% vs (-16.58±1.68) %; global circumferential strain:(-12.13±4.10)% vs (-19±2.24)%; global radial strain:(-7.57±13.55)% vs (-16.58±9.15)%, p<0.001) or segmental level which were prominent in mid and apical segmental longitudinal strain(mid longitudinal strain: (-7.27±3.77)% vs (-21.39±2.72)%; apical longitudinal strain: (-10.08±4.12)% vs (-18.34±2.85)%, p<0.001). Significant differences between IVNC, HCM and hypertrophic VNC were only shown in global circumferential strain( (-16.95±2.49)% vs (-17.65±3.91)% vs (-12.13±4.10)%,p<0.01) and apical circumferential strain: (-19.12±3.44)% vs (-20.3±3.88)% vs (-13.16±5.13)%, p<0.01). Whereas, no evident difference were found in other strain parameters between IVNC, HCM and hypertrophic VNC patients, so as to normal controls.Discussion
In VNC patients associated with HCM, EF may not be a sensitive marker reflecting differences between IVNC and HCM, and it is even not sensitive to distinguish them from normal controls. This may undermine the understanding of pathophysiology underlying these subgroups, and what worse is that this may further affect clinical intervention. Strain parameters derived from cine images showed significant differences between groups, which can offer another view point of ventricular wall motion to analyze the development of these disease entities.Conclusion
Hypertrophic VNC has a remarkable different strain property compared with IVNC and HCM, which may be a breakthrough point to understanding underlying mechanism constituting diverse morphologic traits.Acknowledgements
NoneReferences
[1] Arbustini E, Favalli V, Narula N, et al. Left Ventricular Noncompaction: A Distinct Genetic Cardiomyopathy? [J]. Journal of the American College of Cardiology, 2016, 68(9): 949-66.
[2] Biagini E, Ragni L, Ferlito M, et al. Different Types of Cardiomyopathy Associated With Isolated Ventricular Noncompaction [J]. American Journal of Cardiology, 2006, 98(6): 821-4.
[3]Yuan L, Xie M, Cheng T O, et al. Left ventricular noncompaction associated with hypertrophic cardiomyopathy: echocardiographic diagnosis and genetic analysis of a new pedigree in China [J]. International journal of cardiology, 2014, 174(2): 249-59.
[4]Gutberlet M, Pachowsky M, Grothoff M, et al. Value of MRI derived parameters in the discrimination of familial left ventricular noncompaction (LVNC), DCM and HCM in comparison to healthy volunteers [J]. Journal of Cardiovascular Magnetic Resonance, 2011, 13(Suppl 1): P269.
Figures
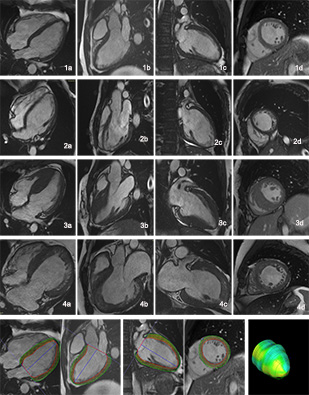
Figure 1: 1-4 represent images from normal control, isolated ventricular noncompaction, hypertrophic cardiomyopathy and hypertrophic ventricular noncompaction respectively with a-d showing different longitudinal and short axial views. the bottom row shows diagram of myocardial strain analysis.