2103
Feasibility Study of Whole Heart Myocardial Perfusion Imaging with tSMS and CS Reconstruction1Shenzhen Institutes of Advanced Technology,Chinese Academy of Sciences, Shenzhen, China, 2Shenzhen College of Advanced Technology, University of Chinese Academy of Sciences, Shenzhen, China, 3UIH America Inc., Houston, TX, United States, 4Shanghai United Imaging Healthcare Co., Ltd, Shanghai, China
Synopsis
Auto-calibrated multiband CAIPIRINHA with through-time encoding (tSMS) has been proposed to acquire multiple slices simultaneously without extra reference scans. Reference images are estimated from the consecutive cardiac phases at a lower temporal resolution for subsequent slice separation. We implemented the tSMS method in a myocardial perfusion sequence and explored the feasibility of whole heart perfusion imaging with tSMS and CS reconstruction retrospectively. The preferable in-vivo results demonstrated the whole heart perfusion imaging is feasible using the tSMS+CS method.
Introduction
First-pass myocardial perfusion MRI shows great potential for ischemia testing in patients with intermediate risk of coronary artery disease[1]. In clinical routine, three equidistant short-axis slices along the LV and a 4-chamber slice are typically acquired over a single heartbeat to cover the entire heart. To date, 3D techniques have been proposed to achieve whole heart coverage[2], which usually reduces in-plane spatial resolution, lengthens the acquisition time within the cardiac cycle and is more susceptible to respiratory motion. Simultaneous multi-slice imaging[3] is an alternative way to increase ventricular coverage without sacrificing in-plane spatial resolution. Recent development in simultaneous multi-slice with through-time encoding (tSMS)[4] allows the simultaneous acquisition and reconstruction of multiple slices in one single breath hold (BH), without the need for extra reference data acquisition, in which separated single-band (SB) reference data is estimated from the consecutive cardiac phases at a lower temporal resolution for subsequent slice separation. The method has been successfully integrated into balance SSFP sequence in our another work[5], which further accelerates the in-plane acquisition using compress sensing (CS). Based on these works, we extend the similar idea into myocardial perfusion imaging and exploit its feasibility of speeding up the acceleration both in in-plane and through plane directions, which enables whole heart coverage within one scan.Method
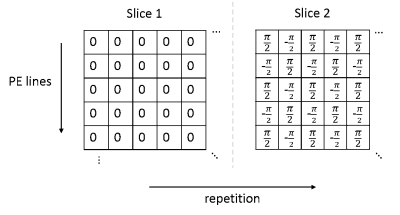
Sequence developmentThe proposed method was implemented into a standard spoiled GRE sequence, with a non-selective saturation recovery (SR) pulse before each shot. The RF phases of the MB pulses of each excited slice are cycled in time (or repetition) as shown in Figure 1. MB=2 was achieved and demonstrated in this study. In-plane CS sampling pattern was generated by the pseudo random in-plane sampling pattern (Figure 2a) using the Latin Hypercube method[6]. Patterns in repetition are different and complementary for adjacent repetitions.
Image acquisition
A healthy subject was imaged on a 3T scanner (uMR790, United Imaging Healthcare, Shanghai, China). A 12-channel abdomen coil in conjunction with a 16-element posterior spine coil was used for data reception. The study was approved by our Institutional Reviews Board (IRB). No contrast agent was administered in the study. Imaging parameters were: FA=15°, TR/TE=2.93/1.41ms, saturation recovery time=100ms, FOV=370×340×10mm3, in-plane matrix size=200×184, spatial resolution=1.85×1.85×10mm3 and bandwidth=1050Hz/pixel. The total scan time was about 50s. The proposed method was retrospectively demonstrated and evaluated using the data acquired from a healthy subject without contrast injected. Data was undersampled according to the k-t acceleration mask as shown in Figure 2b. The undersampled mask was produced by the aforementioned CS sampling pattern.9-fold acceleration in total was achieved with in-plane undersampling factor about 4.5 (40 lines per repetition).
Reconstruction
SB reference data calculation[4]:
Consecutive repetitions can be grouped to generate slice-separated reference datasets at low temporal resolution. Specifically, the aliased k-space data was firstly averaged according to the odd and even cardiac cycles. After that, the SB reference data was calculated by summation and subtraction of the averaged odd and even datasets. Then the separated SB images were reconstructed by Fourier transform and used to estimate coil sensitivity maps for subsequent tSMS+CS reconstruction.
tSMS + CS reconstruction[5]:
Perfusion images were then reconstructed by minimizing cost function below:
$$arg \min_{x_1,x_2} \frac{1}{2}\parallel y-(p_1DF(s_1x_1)+p_2DF(s_2x_2))\parallel _{2}^{2} +\lambda\parallel Tx_1\parallel _1+\lambda\parallel Tx_2\parallel_1$$
where y is the measured data, x1 and x2 are the two perfusion slice groups in time, s1 and s2 are coil sensitivity maps of the SB reference image, p1 and p2 are phase modulations, D represents the k-space sampling operator, F represents the Fourier transform operator, and T represents the temporal TV operator which is the sparsifying transform for L1 regularization, λ is an adjustable parameter of the regularization strength.
Results
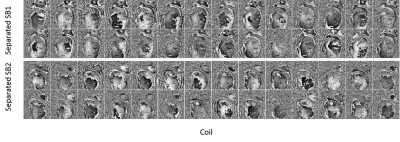
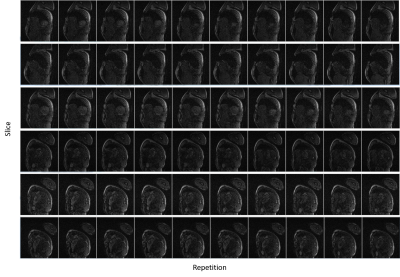
Figure 3 shows a preferable built-in reference image successfully calculated from a highly undersampled data with low temporal resolution. Figure 4 shows coil sensitivity maps of the separated SB images from one MB pair. Some Gibbs artefacts were shown in some coils, which might affect the subsequent reconstruction. An example of separated slices from MB datasets using the proposed tSMS+CS reconstruction was shown in Figure 5. SNR was relatively poor near the myocardium without contrast enhancement, and the signal of myocardium was suppressed due to the saturation pulse.Discussion and Conclusion
One scan, whole heart coverage myocardial perfusion imaging is feasible with the proposed tSMS+CS method, and at least an acceleration of 2 in slice direction can be achieved to coverage the whole heart. Since the in-plane acquisition is highly undersampled, it is able to acquire more slices in the cardiac cycle and has the potential to reduce motion artefact and increase success rates. The in-vivo results are promising and demonstrated the feasibility of whole heart perfusion imaging in one scan using the tSMS+CS method. Potential future directions with the proposed method include sampling pattern optimization, validation in contrasted enhanced perfusion measurement, implementation of whole heart quantitative perfusion imaging, and evaluation of reproducibility.Acknowledgements
Some of the work was partially supported by the National Natural Science Foundation of China (No. 81801691), the State Key Program of National Natural Science Foundation of China (Grant No. 81830056), and the Shenzhen Key Laboratory of Ultrasound Imaging and Therapy (ZDSYS20180206180631473).References
1. Coelhofilho O R, Rickers C, Kwong R Y, et al. Radiology, 2013, 266(3): 701-715.
2. Fair M, Gatehouse P D, Dibella E V, et al. Journal of Cardiovascular Magnetic Resonance, 2015, 17(1): 68-68.
3. Barth M, Breuer F A, Koopmans P J, et al. Magnetic Resonance in Medicine, 2016, 75(1): 63-81.
4. Ferrazzi G, Bassenge J P, Wink C, et al. Magnetic Resonance in Medicine, 2019, 81(2): 1016-1030.
5. Yu Ding, Lele Zhao, et al. Highly Accelerated Free Breathing Whole Heart Real-time Cine using AuTO-calibrated Multiband Imaging and Compressed Sensing (ATOMICS), submitted to ISMRM 2020:2153
6. Lyu, JY, et al, ISMRM 2019, 4752.
Figures