2069
Multiphase CINE T2*-mapping of the mouse heart at 9.4 tesla1Berlin Ultrahigh Field Facility (B.U.F.F.), Max Delbrück Center for Molecular Medicine in the Helmholtz Association, Berlin, Germany, 2Technische Universität Berlin, Berlin, Germany, 3DZHK (German Centre for Cardiovascular Research), partner site Berlin, Germany, 4Experimental and Clinical Research Center, Charite Medical Faculty and the Max Delbrueck Center for Molecular Medicine in the Helmholtz Association, Berlin, Germany
Synopsis
Hypertrophic cardiomyopathy (HCM) is the most frequent inherited monogenic heart disease and could lead to heart failure or even sudden cardiac death. Effective relaxation time T2* is related to different physiological parameters. Dynamic CINE mapping of T2* covering the entire cardiac cycle facilitates distinction of healthy and pathologic myocardial tissue. Here we propose, implement, evaluate and apply an effective approach of retrospective cardiac gating that affords for the first time CINE T2*-mapping in mice at 9.4T and provides the technological basis for translational research into a deeper understanding of the mechanisms underlying hypertrophic cardiomyopathy.
Introduction
Hypertrophic cardiomyopathy (HCM) is the most frequent inherited monogenic heart disease and develops via a progressive process of myocardial remodeling, which eventually leads to heart failure or even sudden cardiac death. Mapping of the effective relaxation time T2* is of great relevance to probe cardiac histopathology since its related to physiological parameters like tissue oxygenation1, blood volume and hematocrit2. Periodical changes of myocardial T2* over the cardiac cycle were reported3 and dynamic T2*-mapping of myocardium across the whole cardiac cycle facilitate distinction of healthy and pathologic tissue in HCM patients5. The relationship between genetic, non-genetic factors, HCM phenotypes and clinical outcomes remains poorly understood and is not trivial to evaluate in human subjects. Here we suggest using mouse models to provide a deeper understanding of underlying mechanisms of HCM. To meet this goal we developed an efficient cardiac retrospective gating approach to permit multiphase T2* weighted imaging and T2*-mapping of the mouse heart covering the entire cardiac cycle.Method
Data acquisition: In vivo cardiac MRI was performed in C57BL/6J mice (average heart rate:= 400-600 bpm) using a 9.4T animal MR scanner (Bruker, Germany). For T2*-mapping multi-, gradient-echo imaging was conducted (TE/TR=2.1-11.9/20ms, flip angle=10°, FOV=(35x35)mm2, matrix=128x128, slice thickness=0.8mm). Data acquisition was repeated continuously for 14 minutes, yielding 300 k-spaces. During MRI a pulse oximetry based TTL trigger signal (Model 1025, SA Instruments, USA) and a TTL signal generated at the start of each k-space acquisition (added to pulse sequence) were recorded simultaneously using an analogue-digital converter (DT 9800-16SE-BNC, Data Translation GmbH, Gemany) and dedicated data acquisition software (HAEMODYN, Hugo Sachs Elektronik – Harvard Apparatus GmbH, Germany).Image reconstruction: Pulse oximetry based retrospective gating was implemented in MATLAB R2017b (The MathWorks, Inc., USA). Based on recorded trigger time points the time window of each heart beat was calculated and divided into cardiac phases. The acquired k-spaces were then assigned to the corresponding phases of the cardiac cycle.
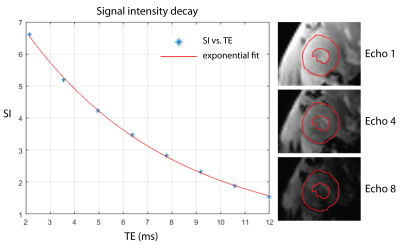
Analysis: T2*-maps of the myocardium were calculated using a mono-exponential signal decay model. The myocardium was manually segmented for each cardiac phase. Quantitative analysis was performed in the septal regions since it has been shown to provide most reliable T2* values due to the immunity to macroscopic susceptibility gradients4.
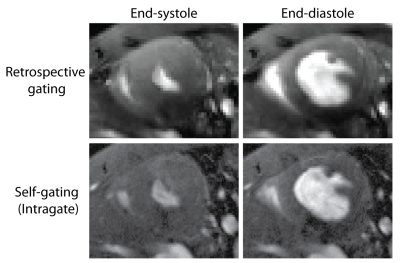
Validation: For demonstrating the feasibility of our retrospective gating approach we applied it to multi-phase short axis views of the mouse heart (FLASH, TE/TR=2.1/10ms, flip angle=20°, FOV=(20x20)mm2, matrix=192x192, slice thickness=0.8mm) which we compared to FLASH images acquired in the same animal using an established self-gating approach (IntraGate, Bruker Biospin, Germany).
Results
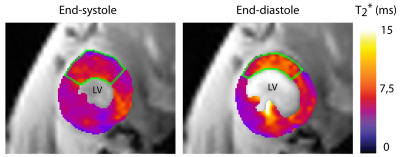
After applying retrospective gating to FLASH technique, images showed overall sharpness which is sufficient to capture cardiac dynamics with a minimum of ghosting and reconstruction artifacts (Fig. 1). Image quality obtained with our retrospective gating approach was found to be comparable to the established self-gating approach (Fig. 1). Retrospective gating permitted dynamic T2*-mapping of the rapidly beating mouse heart covering the entire cardiac cycle (number of cardiac phases=7). Left ventricular myocardium showed the expected exponential signal decay (Fig. 2), allowing accurate calculation of T2*. Mean septal T2* was found to be = (7.37 ± 1.35) ms at end-systole and (9.23 ± 1.65) ms at end-diastole (Fig. 3), showing a T2* increase during the diastolic phase compared to the systolic phase, which agrees with earlier reports in healthy volunteers5.Conclusion and Discussion
In this work we demonstrated the feasibility of retrospective cardiac gating for CINE T2*-mapping of the mouse heart at 9.4T. This retrospective cardiac gating implementation for multi-phase T2* mapping contributes to the technological basis for myocardial T2* monitoring across the cardiac cycle. It can take into account variations in heart rates throughout the scan, and permits rejection of mistriggered data acquisitions. It can be further extended to work directly with the ECG trace, which could improve gating when the ECG is distorted due to gradient-switching and magneto-hydrodynamic effects. This flexible gating approach is compatible with various pulse sequences and MRI protocols and will be further applied to HCM mouse models with the ultimate goal to provide a deeper understanding of the mechanisms underlying HCM while balancing our previous clinical studies with experimental and translational work.Acknowledgements
For the competent and comprehensive support in all scientific matters and also for the open and cordial working atmosphere, I would like to thank the whole B.U.F.F. team. Especially I thank Min-Chi Ku, Andreas Pohlmann and Joao dos Santos Periquito for all your advice and help, which made this work possible. Many thanks also to Prof. Thoralf Niendorf, who provided me the opportunity to be part of the team and lead me to great scientific work with beneficial and comprehensive support.References
1. Friedrich et al. (2013) J Cardiovasc Magn Reson 15:43.
2. Yablonskiy et al. (1994) Magnetic Resonance in Medicine 32(6):749.
3. Huelnhagen et al. (2017) Magnetic Resonance in Medicine 77(6):2381.
4. Meloni et al. (2014) Magn Reson Med 71(6):2224. 5. Huelnhagen et al. (2018) Sci Rep 8:3974.
Figures