2013
U-net based automatic segmentation of the vocal tract airspace in speech MRI1Biomedical Engineering, University of Iowa, iowa city, IA, United States
Synopsis
We develop a fully automated airway segmentation method to segment the vocal tract airway from surrounding soft tissue in speech MRI. We train a U-net architecture to learn the end to end mapping between a mid-sagittal image, and the manually segmented airway. We base our training on MRI of sustained speech sound database and dynamic speech MRI database. Once trained, our model performs fast airway segmentations on unseen images. We demonstrate the proposed U-NET based airway segmentation to provide considerably improved DICE similarity compared to existing seed-growing segmentation, and minor differences in DICE similarity compared to manual segmentation
Purpose
Speech production involves a complex and intricate coordination of several articulators such as the lips, tongue, velum, pharyngeal wall, glottis, and epiglottis. As these articulators move, the vocal tract airway geometry deforms and undertakes specific poses for specific sounds. Segmenting the vocal tract airway geometry, therefore, forms a crucial step in characterizing the underlying vocal tract airspace. Current segmentation algorithms in speech MRI range from segmenting the articulators themselves [1], segmenting the air-tissue interfaces [2], or segmenting the vocal tract air-space [3]. In this work, we develop a novel fully automatic rapid vocal air-space segmentation algorithm based on the U-NET architecture.Methods
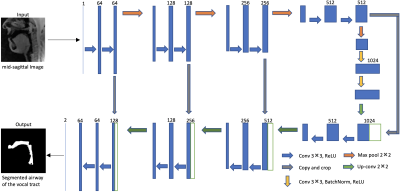
We employ the U-net architecture [4] to learn the mapping of a mid-sagittal speech image at the input and airway manual segmentation at the output (see Figure 1). We have used two sources of speech MRI databases. The first source was the University of Southern California’s (USC) speech morphology database which contained volumetric scans of the airway during sustained production of vowel and consonant sounds [13]. We considered only the 2D problem of segmenting the airway from the mid-sagittal section of these volumetric scans. The second source was from the dynamic 2D mid-sagittal sequence acquired with a GRE sequence (spatial resolution: 2.7 mm2; ~6 frames/second, FOV: 20x20 cm2). Two subjects were scanned with this dynamic sequence at our host institution and were instructed to speak a range of speech stimuli in a slow and controlled pace to avoid motion artifacts. These stimuli included productions of sounds, za-na-za, zi-ni-zi,loo-lee-laa, apa-ipi-upu, counting numbers, and speaking a spontaneous sentence. We trained the U-Net independently with these two databases using a total of 75 images from each of the databases. Another split of data included 5 images for validation; and 20 images for testing. Relevant training parameters were number of epochs = 100, batch size = 2, learning rate =0.0001, dropout rate = 0.5, and a choice of the adaptive moment estimation (ADAM) optimizer. The total training time was 20 hours. The architecture was implemented in Keras with TensorFlow backend on an Intel Core-i7 8700CK, 3.70 GHz 12 core CPU machine. For the sustained speech sound database, we compared the performance of U-net against existing seed-growing segmentation [5] and manual segmentation. Manual segmentation obtained from user-1 was considered as the reference mask. The performances were evaluated in terms of DICE similarity (D) of the estimated segmentation mask with the reference mask. For the second database (dynamic speech), we compared U-net against manual segmentation from user-2 using manual segmentation from user-1 as a reference. Seed-growing was not implemented in the dynamic database because of non-trivialness in defining multiple disconnected regions and specifying seeds in these regions.Results
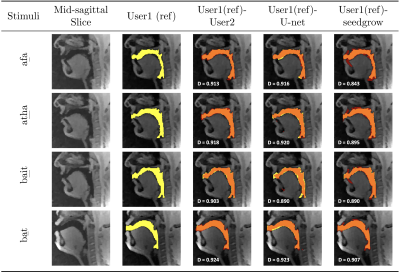
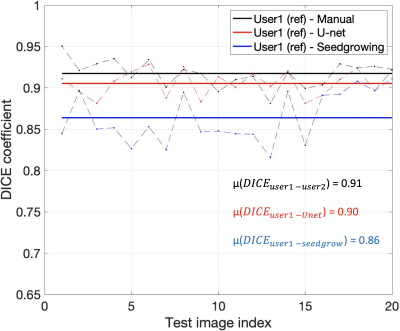
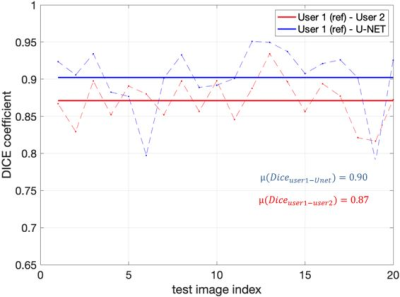
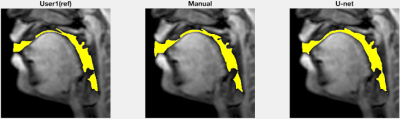
Figure 2 shows representative segmentations of consonant sounds (atha, afa), and vowel sounds (bait, bat) from the test set of the sustained speech sound database. The reference segmentations from user-1 are overlaid with the segmentations from the three schemes of the manual segmentation from user-2, proposed U-net segmentation, and the seed-growing segmentation. The DICE coefficients are shown on the images. While seed-growing segments the vocal airspace, it is sensitive to leaking into airspaces beyond the vocal tract. In contrast, U-net consistently provided a higher DICE similarity, and depicted good segmentation accuracy, and has similarities to the manual segmentations. Figure 3 shows the average DICE similarity of all the methods with reference segmentations on all the 20 test images on the sustained speech sound database. The U-net segmentation (mean DICE = 0.9) shows a considerable improvement over seed-growing (mean DICE = 0.86), and a minor difference compared to a manual segmentation from user-2 (mean DICE = 0.91). Figure 4 similarly shows the average DICE the similarity of all methods on the 20 test images on the dynamic MRI database. The test images correspond to a speech stimulus of spontaneous speech where the vocal airspace is deforming continuously with time. Figure 5 finally shows the animations of the segmentations on the dynamic MRI database. On all the test images, U-net also demonstrated faster processing times to generate the segmentation. The average mean processing times were 0.21sec/image for U-net, 11.6sec/image for seed-growing and 45 sec/image for the manual segmentations.Conclusion
We successfully demonstrated airway segmentation of the vocal tract in speech MRI using the U-NET architecture. The architecture is trained to learn the end to end mapping between the mid-sagittal image and the segmented airway. Once trained, the model performs fast airway segmentations at the order of 0.21s per slice on a modern CPU with 12 cores. We demonstrated this on two databases (sustained speech database, and a dynamic speech database). With U-net, we showed improved DICE similarity compared to existing seed-growing segmentation, and minor differences in DICE similarity compared to manual segmentation.Acknowledgements
No acknowledgement found.References
[1] Z. I. Skordilis, V. Ramanarayanan, L. Goldstein, and S. S. Narayanan, “Experimental assessment of the tongue incompressibility hypothesis during speech production,” in Proceedings of the Annual Conference of the International Speech Communication Association, INTERSPEECH, 2015.
[2] E. Bresch and S. Narayanan, “Region segmentation in the frequency domain applied to upper airway real-time magnetic resonance images,” IEEE Trans. Med. Imaging, 2009.
[3] K. Somandepalli, A. Toutios, and S. S. Narayanan, “Semantic edge detection for tracking vocal tract air-Tissue boundaries in real-Time magnetic resonance images,” in Proceedings of the Annual Conference of the International Speech
[4] O. Ronneberger, P. Fischer, and T. Brox, “U-net: Convolutional networks for biomedical image segmentation,” in Lecture Notes in Computer [5] Z. I. Skordilis, A. Toutios, J. Toger, and S. Narayanan, “Estimation of vocal tract area function from volumetric Magnetic Resonance Imaging,” in ICASSP, IEEE International Conference on Acoustics, Speech and Signal Processing - Proceedings, 2017.
Figures