1922
Machine-Learning-Based Segmentation of Ischemic Penumbra By Using Diffusion Tensor Metrics in a Rat Model1Department of Medical Imaging, Taipei Medical University Hospital, Taipei, Taiwan, 2Translational Imaging Research Center, Taipei, Taiwan, 3Department of Radiology, School of Medicine, College of Medicine, Taipei Medical University, Taipei, Taiwan, 4Radiogenomic Research Center, Taipei Medical University Hospital, Taipei, Taiwan, 5Institute of Statistical Science, Academia Sinica, Taipei, Taiwan, 6TMU Research Center for Artifical Intelligence in Medicine, Taipei, Taiwan, 7Graduate Institute of Biomedical Electrics and Bioinformatics, National Taiwan University, Taipei, Taiwan
Synopsis
In the present study, we developed a 2-level classification model with an overall accuracy of 88.1 ± 6.7% for discriminating the stroke hemisphere into the infarct core (IC), ischemic penumbra (IP), and normal tissue regions on a voxel-wise basis in a permanent left middle cerebral artery occlusion model. According to the analysis results, we suggest that a single diffusion tensor imaging (DTI) sequence combined with machine learning (ML) algorithms can dichotomize ischemic tissue into the IC and IP, which are comparable to the conventional perfusion–diffusion mismatch.
Background and Purpose:
Recent trials have shown promise in intra-arterial thrombectomy after the first 6-24 hours of stroke onset. Quick and precise identification of the salvageable tissue is essential for successful stroke management. In this study, we examined the feasibility of machine learning (ML) approaches for differentiating the ischemic penumbra (IP) from the infarct core (IC) by using diffusion tensor imaging (DTI)-derived metrics.Methods:
This study was approved by the local institutional animal care and use committee. Fourteen male rats subjected to permanent middle cerebral artery occlusion (pMCAO) were included for analyses. Using a 7T magnetic resonance imaging, DTI metrics such as fractional anisotropy(FA), pure anisotropy(q), diffusion magnitude(L), mean diffusivity (MD), axial diffusivity(AD), and radial diffusivity(RD) were derived. The MD and relative cerebral blood flow maps were coregistered to define the IP and IC at 0.5 hours after pMCAO(figure 1A). Three types of features (the 6 relative DTI-derived metrics(figure 1B), Mahalanobis distance, and normalized histogram along with its kurtosis and skewness) were extracted from regions of interest in the voxel-located slices and adjacent slices. A 2-level classifier was proposed based on DTI-derived metrics to classify stroke hemispheres into the IP, IC, and normal tissue (NT). In this study, support vector machine (SVM) with a cubic kernel1 was used as a classification algorithm2 and the classification performance was evaluated through the leave-one-out cross validation method, which were implemented using the Statistics and Machine Learning Toolbox in the MATLAB environment.Results:
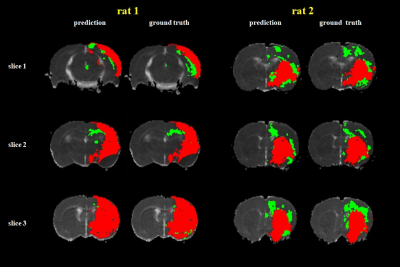
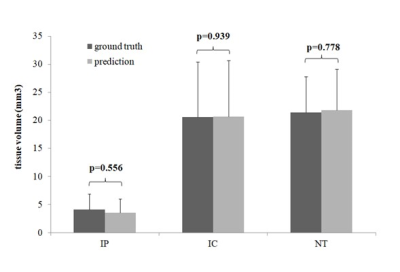
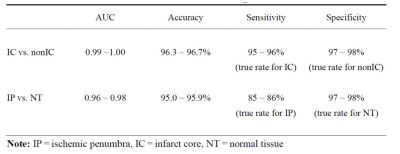
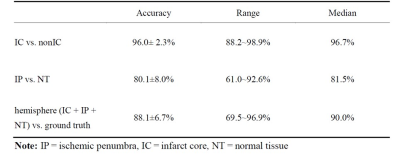
The IC and non-IC can be accurately segmented by the proposed 2-level classifier with an area under the curve (AUC) between 0.99 and 1.00 and accuracies between 96.3 and 96.7%. For the training dataset(Table 1), the non-IC can be further classified into the IP and NT with an AUC between 0.96 and 0.98 and accuracies between 95.0 and 95.9%. For the testing dataset(Table 2), the classification accuracies between the IC and non-IC and between the IP and NT were 96.0 ± 2.3% and 80.1 ± 8.0%, respectively. The accuracy of the segmentation for 3 tissue subtypes in the stroke hemisphere was 88.1 ± 6.7%. Figure 2 illustrates the comparison of the classifier-defined IC and IP with the corresponding perfusion–diffusion-defined IC and IP for a rat. In the suture-occlusion model, the IP is relatively small (even sparse) in areas at the margin of a large IC. Nevertheless, the proposed classification model can successfully segment the ischemic regions into the IP and IC through visual inspection. Moreover, the lesion volumes predicted by the proposed classifiers were not significantly different from those of the ground truth for the 3 tissue subtypes (P = .56, .94, and .78, figure 3).Conclusions:
A single DTI sequence along with ML algorithms can dichotomize ischemic tissue into the IC and IP, which are comparable to conventional perfusion–diffusion mismatch.Acknowledgements
No acknowledgement found.References
1.Schölkopf B, Smola AJ, Bach F. Learning with kernels: Support vector machines, regularization, optimization, and beyond. MIT press; 2002.
2. Prajapati GL, Patle A. On performing classification using svm with radial basis and polynomial kernel functions. 2010 3rd International Conference on Emerging Trends in Engineering and Technology. 2010:512-515
Figures
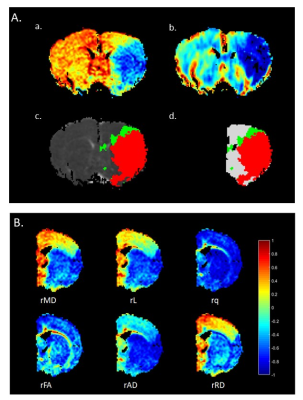
Figure 1A. Definitions of the IP, IC, and NT in a Rat Subjected to pMCAO. IC was defined as the blue area in the MD map (a) and perfusion deficit is shown in (b). Perfusion–diffusion mismatch is illustrated in (c) and (d), where the red region indicates the IC and the green region indicates the IP. The NT was defined as the region in the ipsilateral hemisphere except for the IP and IC [white in (d)]. Figure 1B. Maps of Relative DTI Metrics. The intensity of the map represents the quantitative decreases or increases of the DTI metrics compared with the corresponding contralateral homologous tissue.