1912
Validating multimodal MRI based stratification of IDH genotype using radiomics and CNNs
Madhura Ingalhalikar1, Tanay Chougule1, Sumeet Shinde1, Vani Santosh2, and Jitender Saini3
1Symbiosis Center for Medical Image Analysis, Symbiosis International University, Pune, India, 2Department of Neuropathology, National Institute of Mental Health and Neurosciences, Bangalore, India, 3Department of Radiology, National Institute of Mental Health and Neurosciences, Bangalore, India
1Symbiosis Center for Medical Image Analysis, Symbiosis International University, Pune, India, 2Department of Neuropathology, National Institute of Mental Health and Neurosciences, Bangalore, India, 3Department of Radiology, National Institute of Mental Health and Neurosciences, Bangalore, India
Synopsis
Radiomics based multi-variate models and state-of-art convolutional neural networks (CNNs) have demonstrated their usefulness for predicting IDH genotype in gliomas from MRI images. However, adaptability and clinical explanability of these models on unseen multi-center datasets has not been investigated. Our work trains radiomics and CNN based classifiers on a large dataset (TCIA) and tests multiple local datasets. Results demonstrate higher adaptability of radiomics than standard CNNs, except for transfer learned CNNs. Better interpretability was obtained from feature ranking (in case of radiomics) and high resolution class activation maps (in case of CNNs).
Introduction
Mutations in isocitrate dehydrogenase 1(IDH1) in diffuse gliomas have been considered crucial as these are associated with longer overall survival1. Currently, the IDH genotype is identified via immuno-histochemical analysis following biopsy or surgical resection. Therefore, developing non-invasive pre-operative markers for IDH genotype is clinically important as it can not only aid prognosis but also support treatment planning and therapeutic intervention. Currently (1) radiomics from multi-parametric MRI with multi-variate classification and (2) deep learning technique of convolutional neural nets (CNN) have increasingly gained attention and has already been applied for the identification of the IDH mutation status2,3,4,5. However, these studies do not perform leave-one-site-out type of analysis creating uncertainties about the adaptability to unseen datasets acquired from different scanners with diverse scanning protocols. Moreover, CNN based methods, although illustrate higher discriminative power, do not provide insights into the regions or features that discriminate one class from another which is crucial for clinical interpretability. To mitigate the aforementioned issues, this work compares radiomics based and CNN classifier trained on a large open source dataset and tests it on locally acquired datasets to assess the applicability of these models. For clinical interpretability, we extract the underlying discriminative radiomics features and for the CNN model we employ a high resolution class activation map (HR-CAM) technique to demonstrate the regions of discrimination on multiple modalities.Methods
Data from TCIA repository which included 90 subjects with IDH mutation and 57 wildtype that were pre-processed and segmented as given in Bakas et al.6 was used for training and cross-validation. Our local datasets consisted of clinical cohort(s) of subjects that had undergone surgical resection, standard post-surgical care and were identified retrospectively after reviewing the medical records. IDH mutation status was determined via immuno-histochemistry or next generation sequencing. The demographic and clinical information is provided in Table 1. Cohort 1 was scanned on a Philips Achieva 3T MRI scanner where (1) T1 weighted (T1ce) :TR/TE=8.7/3.1 ms using a TFE sequence (2) FLAIR: TR/TE/T1=11000/125/2800 ms, in plane resolution = 0.5x0.5mm (3) T2: TR/TE=3600/80 ms and 0.5*0.5 mm resolution in the axial plane. For cohort 2, (1) T1ce: TR/TE/TI=2200/2.3/0.9 ms, T1 MPRAGE sequence with 1*1*1 mm isotropic resolution (2) FLAIR: same as cohort 1 (2) T2: TR/TE ranging from 5500/90ms and 0.5*0.5 mm resolution in the axial plane. Preprocessing included brain extraction, inhomogeneity correction7 and intensity normalization followed by tumor segmentation that was performed using an auto-encoder and later corrected manually. The TCIA/TCGA data was divided into training cohort (74 Mutant, 41 WT) and validation cohort (16 Mutant, 16 WT) and the other two datasets were used for testing. Radiomics: Feature extraction was performed using PyRadiomics 2.2.0 library8 and included statistical features and multiple textural features. A total of 321 features overall (for 3 modalities) were computed and used in a random forest classifier (RF Classifier). CNNs: A CNN with high resolution class activation maps9 architecture (Fig. 1) was employed on a boxed region around tumor for each 2D-axial slice. To perform transfer learning, weights learned from the TCIA dataset were used as initial weights for the CNN and it was then re-trained on the combined test datasets for 100 epochs. Here we combined test cohort 1 and 2 for training and testing where 48 subjects were used for transfer learning and 16 were used for testing.Results
CNNs and Radiomics were compared using 5-fold cross-validation on TCIA dataset. The performance of CNNs was better with 95.3% accuracy (Sensitivity/Specificity:0.96/0.94) compared to radiomics with 86.9% accuracy (Sensitivity/Specificity:0.87/0.85). Whereas, for unseen test data, we observed that radiomics performed with a higher accuracy (67.5% and 83.3%) while CNN model demonstrated lower accuracy(67.5% and 70.1%). However, with transfer learning we could improve the performance of CNNs to 81%. Fig. 2 demonstrates ROC curves for all three datasets. Fig. 3 illustrates the top features obtained from the random-forest model. Finally, Fig. 4 provides an example of the HR-CAMs, that illustrate the most discriminative region for each subject under consideration. The red area on the HR-CAMs is highly weighted by the CNNs.Conclusion
Our results demonstrated that although CNNs were better in training and cross-validation, radiomics based classification was more robust on unseen data. However, CNNs with the option of using transfer learning demonstrated a boost in accuracy. Furthermore, we also demonstrated that T1ce and T2 based Radiomics features were significant in delineating IDH genotype. With the CNNs we illustrated that patient specific HR-CAMs can be employed to gain insights into the most discriminative regions that might hold implications in targeted therapy. The findings of this study are crucial as imaging prediction of IDH mutation is important and as and when IDH mutant inhibitors become clinically available, these might be used as neoadjuvant therapy.Acknowledgements
No acknowledgement found.References
- Houillier, C., et al., IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology, 2010. 75(17): p. 1560-6.
- Lu, C.F., et al., Machine Learning-Based Radiomics for Molecular Subtyping of Gliomas. Clin Cancer Res, 2018. 24(18): p. 4429-4436.
- Suh, C.H., et al., Imaging prediction of isocitrate dehydrogenase (IDH) mutation in patients with glioma: a systemic review and meta-analysis. Eur Radiol, 2019. 29(2): p. 745-758.
- Li, Z., et al., Deep Learning based Radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma. Sci Rep, 2017. 7(1): p. 5467.
- Chang, K., et al., Residual Convolutional Neural Network for the Determination of IDH Status in Low- and High-Grade Gliomas from MR Imaging. Clin Cancer Res, 2018. 24(5): p. 1073-1081.
- Bakas, S., et al., Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data, 2017. 4: p. 170117
- N.J Tustison, B.B Avants, P.A. Cook et al., "N4ITK: improved N3 bias correction", IEEE Trans. Med. Imaging, vol. 29, no. 6, pp. 1310-1320, 2010.
- Joost J.M. van Griethuysen et al, “Computational Radiomics System to Decode the Radiographic Phenotype”.
- Shinde S., Chougule T., Saini J., Ingalhalikar M. (2019) HR-CAM: Precise Localization of Pathology Using Multi-level Learning in CNNs. MICCAI 2019. Lecture Notes in Computer Science, vol 11767. Springer, Cham
Figures
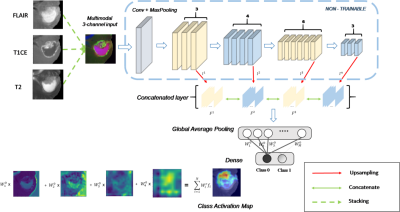
Fig.
1: CNN architecture
employed for predicting the IDH genotype as well as generating
HR-CAMs.
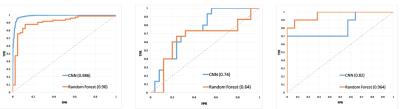
Fig.
2: ROC curves for
(a) CV- on the TCGA/TCIA data (b) Local test dataset 1 and (c) Local
test dataset 2.
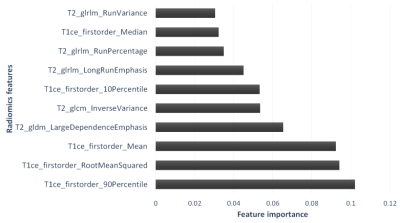
Fig. 3: Top ranked radiomics features plotted against the RF score (Feature importance). It can be observed that the first order features from the enhancing area on T1ce are important in classifying IDH followed by texture features extracted on T2 images.
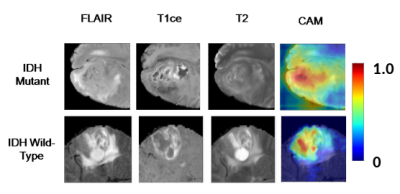
Fig. 4: Figure showing
the HR-CAMs for (row1) –mutant case and (row2) wildtype case. The
first column is the T2-FLAIR, second is the T1ce and third column in
the T2 weighted image. The final column shows the HR-CAM that
illustrates the most discriminative area.
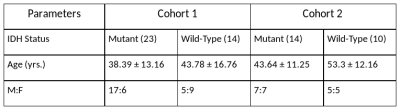
Table 1 : Demographic and clinical Information