1902
Deep Learning for Classifying Patients with Obstructive Sleep Apnea from Healthy Controls using High-resolution T1-weighted Images1Statistics, University of California at Los Angeles, Los Angeles, CA, United States, 2Anesthesiology, University of California at Los Angeles, Los Angeles, CA, United States, 3Medicine, University of California at Los Angeles, Los Angeles, CA, United States, 4Psychiatry and Biobehavioral Sciences, University of California at Los Angeles, Los Angeles, CA, United States, 5Radiology, University of California at Los Angeles, Los Angeles, CA, United States, 6Bioengineering, University of California at Los Angeles, Los Angeles, CA, United States, 7Brain Research Institute, University of California at Los Angeles, Los Angeles, CA, United States
Synopsis
Deep learning has demonstrated impressive performance in a wide range of complex and high-dimensional imaging data, including medical image classification and segmentation. One major challenge of harnessing the power of neural networks in image analysis is the small sample size. The present work utilizes deep learning models to classify high-resolution T1-weighted images of obstructive sleep apnea patients (OSA) from healthy controls. Using 193 participants and with adopted model regularization and exponential moving averaging of model weights, we showed 65% testing accuracy and 80% sensitivity. The findings demonstrate the potential for applying neural network models in assisting image-based OSA diagnoses.
Introduction
Deep learning has demonstrated significant performance in a wide range of complex and high-dimensional data domains, including natural images, 1 as well as medical image classification and segmentation.2 Neural network models have been mathematically proven to be a universal function approximator, and are able to estimate the complex functional relations among high dimensional data, such as medical imaging. Strong model expressivity is a major advantage of deep learning over traditional machine learning approaches. Although traditional methods require hand-crafted features, the flexible deep learning models can learn features directly from imaging data, which reduces the demand of domain-specific knowledge to design features and improves the model performance. Despite the advantage, there is a major challenge of harnessing the power of neural networks in image analysis, which requires a significantly large sample size. However, in imaging studies and applications, the sample size is typically small, varying from less than one hundred to a few hundred samples. The present work utilizes deep learning models to classify high-resolution T1-weighted images of obstructive sleep apnea (OSA), a condition characterized by successive collapses of the upper airway muscle with continued diaphragmatic movement to breathe during sleep, resulting to multiple hypoxic/ischemic episodes every night, from those with healthy controls using the three-dimensional (3D) Convolutional Neural Network (CNN) models. We adopted model regularization and exponential moving averaging of model weights to address the issue of small sample size and improve model performance.Materials and Methods
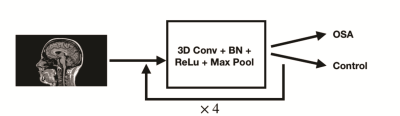
One hundred ninety-three participants were studied. Of 193, 79 participants were diagnosed with OSA [(mean± SD), age, 49.3±9.9 years; BMI, 32.2±6.6 kg/m2; apnea-hypopnea-index, 37.7±22.9 events/hour; 48 males] and 114 participants (age, 49.2±9.5 years; BMI, 24.1±3.6 kg/m2; 44 males) were healthy controls. Brain MRI scans were acquired using a 3.0-Tesla MRI scanner (Siemens, Magnetom, Prisma). Two high-resolution T1-weighted images were acquired using a magnetization-prepared rapid gradient-echo sequence (MPRAGE) pulse sequence (TR = 2200 ms; TE = 2.4 ms; inversion time = 900 ms; flip angle (FA) = 9°; matrix size = 320×320; field of view (FOV) = 230×230 mm2; slice thickness = 0.9 mm; number of slices = 192) from all the subjects. Both image series were realigned and averaged to improve SNR. We employed 69 OSA and 104 healthy control subjects for model training and validation, and 10 OSA and 10 healthy control subjects were used for testing the model. For the training data, OSA group was up-sampled to match the number of samples in the control group. We designed a 3D CNN model to classify OSA patients from healthy controls, which is shown in Figure 1. Due to the small sample size, L2 regularization was applied to the model to minimize overfitting. Also, we kept an exponential moving average (EMA) of model weights, while training the model and the EMA of weights were used in classifying testing data. Thus, the model weights were less heavily affected by the local data, mitigating overfitting.Results
We observed a testing accuracy of 65% in classifying MRI images from OSA and controls. The sensitivity of model was 80%, indicating that there is a 80% chance for a patient to be diagnosed as OSA, given subject indeed has the OSA condition. These results suggest that method has a low likelihood to misdiagnose those subjects with OSA. The specificity of model was 50%, indicating that a healthy person has a chance level to be diagnosed with OSA.Discussion
OSA subjects showed microstructural changes in multiple brain sites. However, high-resolution T1-weighted conventional images did not show significant differences between OSA and healthy control subjects. Despite the small sample size, the modeling techniques adopted in this work, model regularization and EMA, can control overfitting and result in a reasonable classification accuracy of high-resolution T1-weighted data and sensitivity of diagnosis. The findings indicate that deep learning methods can be used in the classification of OSA from healthy controls, which might aid in the future diagnosis of the condition without a traditional diagnosis.Conclusion
The findings demonstrate the potential of applying deep learning models to assist image-based diagnoses of OSA.Acknowledgements
This work was supported by NIH R01 NR-015038.References
LeCun, Y., Bengio, Y. & Hinton, G. Nature 521, 436–444 (2015).
Kermany, D. S. et al. Cell 172, 1122–1131 (2018).