1895
Deep Learning synthesis of PET-like CBF maps from arterial spin labeling MRI: Rapid Clinical Deployment & Evaluation
David D Shin1, David Yen-Ting Chen2, Audrey P Fan2, Moss Y Zhao2, Jia Guo3, Suchandrima Banerjee1, and Greg Zaharchuk2
1GE Healthcare, Menlo Park, CA, United States, 2Stanford University, Stanford, CA, United States, 3University of California, Riverside, CA, United States
1GE Healthcare, Menlo Park, CA, United States, 2Stanford University, Stanford, CA, United States, 3University of California, Riverside, CA, United States
Synopsis
Deep learning ASL Pipeline is presented that automatically retrieves relevant MR images from multiple clinical scanners, performs preprocessing/inferencing and returns the derived synthesized PET-like CBF maps back to the scanners for rapid clinical evaluation and prototyping.
INTRODUCTION
Arterial spin labeling (ASL) has become a routine MR imaging technique for quantitative measure of cerebral blood flow (CBF) (1). However, one main limitation remains in its sensitivity to transit delay (TD) effects. In clinical cases where the TD is long (e.g. ischemic stroke, Moyaomoya), perfusion of the affected region can be severely underestimated (2,3). While there are alternate techniques to mitigate this effect (i.e. Multi-delay TD mapping, velocity selective ASL), both methods present their own challenges (4–6).PET O15 derived CBF is not constrained by these limitations and therefore can serve as an excellent Deep Learning (DL) training data to improve the accuracy and reliability of ASL-derived CBF maps, particularly when the data from both modalities are acquired simultaneously (7). Here we present the DL-ASL Pipeline that automatically retrieves relevant MR images from multiple clinical scanners, performs preprocessing/inferencing and returns the synthesized PET-like CBF maps back to the scanners for rapid clinical evaluation and prototyping.
METHODS
Figure 1 is a schematic of the DL-ASL Pipeline workflow.The source code driving the workflow is self-contained and is hosted on GitHub. The code needs to be installed both on the scanner and on the server where preprocessing and DL inferencing takes place.
The source code resides both on the scanner and a server where the preprocessing and inferencing takes place. Current implementation allows multiple scanners connected to the same processing unit.
In our current implementation, the DL inference requires data from multiple scans, i.e. 1) single-TI PCASL; 2) multi-TI PCASL; 3) MPRAGE T1; and 4) T2 FLAIR. The search for these scans in a session is auto triggered at the end of the Multi-TI PCASL run, using the unique series name corresponding to each scan as defined in the protocol. A user modifiable text file is provided to ensure that each site can use its specific series names as the search criteria.
The pipeline also accommodates a scan session where two sets of ASL scans are performed before and after an intervention (e.g. Diamox, caffeine administration). In this case, the operator can simply acquire two sets of ASL scans separated by the T1 and/or T2 scan. The retrieved DICOM images are anonymized and sent to a GPU server via SCP where the images are co-registered and normalized to MNI space, and where inferencing is performed. The output CBF maps are sent back to the source scanner in DICOM format and appear on the scanner console within the same session from which the original images were sent.
RESULTS
Figure 2 shows the original and inferenced CBF maps from a healthy volunteer and a Moyamoya patient as were generated by the DL-ASL Pipeline and presented to the respective scanner, i.e. a 3T MR750 Discovery and a Signa PET/MR (GE Healthcare, Milwaukee, WI). For each case, the skull-stripped T1W image corresponding to the CBF slice is shown, which is performed during the preprocessing step. Figure 2: CBF maps generated from the DL-ASL Pipeline from two subjects. CBF values are shown in absolute units (ml/100g-min). In both cases, there were two sets of ASL scans, i.e. pre and post Diamox, which the DL-ASL Pipeline recognized and processed successfully.DISCUSSION & CONCLUSION
Current implementation uses a dedicated server as a main processing engine. However, the proposed framework can be hosted in the cloud or on a vendor specific platform (e.g. GE Edison) to achieve high throughput and minimize computation time. In conclusion, the pipeline presented here allows a rapid prototyping of the DL inferenced ASL technique and easy integration to the existing clinical workflows and can be deployed to multiple scanners both within and across institutions.Acknowledgements
No acknowledgement found.References
1. Haller S, Zaharchuk G, Thomas DL, et al. Arterial Spin Labeling Perfusion of the Brain: Emerging Clinical Applications. Radiology. 2016;281(2):337–356. 2. Zaharchuk G. Arterial Spin Labeled Perfusion Imaging in Acute Ischemic Stroke. Stroke. 2014;45(4):1202–1207. 3. Fan AP, Guo J, Khalighi MM, et al. Long-Delay Arterial Spin Labeling Provides More Accurate Cerebral Blood Flow Measurements in Moyamoya Patients: A Simultaneous Positron Emission Tomography/MRI Study. Stroke. 2017;48(9):2441–2449. 4. Buxton RB, Frank LR, Wong EC, et al. A general kinetic model for quantitative perfusion imaging with arterial spin labeling. Magn Reson Med. 1998;40(3):383–396. 5. Bokkers RP, Bremmer JP, van Berckel BN, et al. Arterial spin labeling perfusion MRI at multiple delay times: a correlative study with H215O positron emission tomography in patients with symptomatic carotid artery occlusion. J Cereb Blood Flow Metab. 2010;30(1):222–229. 6. Wong EC, Cronin M, Wu W-C, et al. Velocity-selective arterial spin labeling. Magn Reson Med. 2006;55(6):1334–1341. 7. Guo J, Gong E, Goubran M, et al. Improving Perfusion Image Quality and Quantification Accuracy Using Multi-contrast MRI and Deep Convolutional Neural Networks. 26th annual meeting of the Int Soc Magn Reson Med, Paris France: 310 (2017).Figures
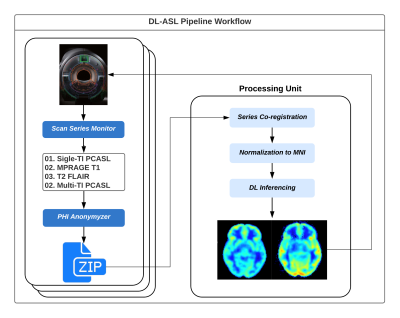
Figure 1: A schematic of the DL-ASL workflow.
The source code resides both on the scanner and a server where the
preprocessing and inferencing takes place. Current implementation allows
multiple scanners connected to the same processing unit.
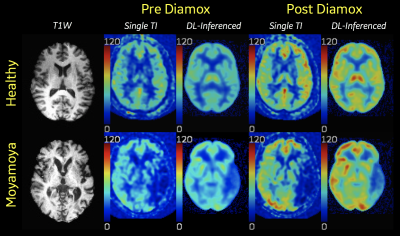
Figure 2 shows the original and inferenced CBF maps from a healthy volunteer and a Moyamoya patient as were generated by the DL-ASL Pipeline and presented[AF1] to the respective scanner, i.e. a 3T MR750 Discovery and a Signa PET/MR (GE Healthcare, Milwaukee, WI). For each case, the skull-stripped T1W image corresponding to the CBF slice is shown, which is performed during the preprocessing step.