1894
Automated Segmentation of Brain Meningioma MRIs with Generative Adversarial Networks1Department of Radiology, University of Cambridge, Cambridge, United Kingdom, 2Department of Neurosurgery, University of Cambridge, Cambridge, United Kingdom
Synopsis
We describe a deep learning method for fully-automated brain meningioma MRI segmentation. A conditional generative adversarial network (cGAN) was trained on T1 contrast-enhanced (T1ce) MRI of 37 patients. We explored the effect of batch size, transfer learning and histogram equalization on segmentation accuracy. The highest results for T1ce images were achieved for meningioma dataset of batch size = 1 (DSC = 0.347). Histogram equalization improved segmentation accuracy for batch size = 1 (DSC = 0.364) and batch size = 200. Transfer learning on a publicly available glioma dataset did not improve segmentation results.
Introduction
Meningiomas are slow-growing tumours arising from arachnoidal cells within meninges covering the brain and spinal cord. Meningiomas are most commonly benign and asymptomatic for many years before they cause symptoms. Magnetic resonance imaging (MRI) is the preferred modality for diagnosis due to its high soft-tissue contrast1.Specific measurements (e.g. tumour volume) are useful for monitoring meningioma growth and treatment planning1. This information is obtained through the laborious and often error-prone process of manual image segmentation. Fully-automated solutions have been developed to improve segmentation efficiency2,3. Deep learning approaches such as convolutional neural networks have been used in meningioma segmentation4 but are limited by the need to determine an effective loss function. The use of two competing neural networks (i.e. generative adversarial networks—GANs) facilitates this process.
We investigate the use of a conditional GAN (cGAN) for automated intracranial meningioma MRI segmentation, and explore the utility of transferring weights determined from Multimodal Brain Tumour Segmentation Challenge (BRATS) 2018 training for improving this task5,6.
Methods
Network architecture: A cGAN7 was used within PyTorch (Torch v0.5, CUDA v9.0) using an NVIDIA Quadro P6000 GPU. The cGAN creates an image through a generator network, and a discriminator network determines if it is ‘real’ or ‘fake’. Images are generated from typically random noise and receives additional image information as input. The “pix2pix” framework8 (a combined U-Net generator and Markov Random Field-like discriminator) was used to generate images indistinguishable from a target image (tumour segmentation). The discriminator performed a patch-wise (64 x 64) classification and then averaged all patches to create a binary output indicating whether the generated image was more ‘fake’ or ‘real’ than noise fed into the generator. To reduce blurring and ensure low-frequency correctness, the L1 distance is incorporated into the loss function of the cGAN8.Datasets:
Meningioma dataset: Contrast-enhanced T1 brain MR images were obtained from 37 patients on 1.5 and 3.0T MRI systems from 2010 to 2018. Lesion masks were segmented manually by trained radiologists.
BRATS 2018 dataset: Contrast-enhanced T1 brain images with segmentation masks were obtained from a publicly available database5,6. MRI for low-grade (n = 75) and high-grade (n = 210) gliomas were used.
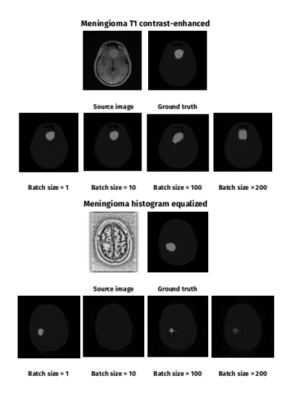
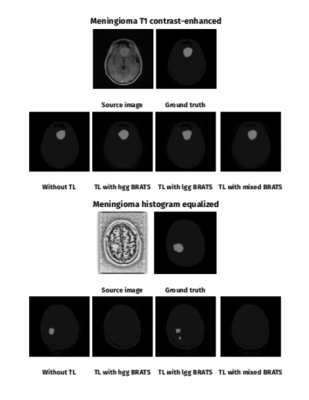
Training: Each network was trained for 100 epochs. Initially, separate trainings were run on the meningioma and BRATS datasets. For BRATS training, networks were trained on low, high and mixed grade MRI. Patients for the mixed BRATS dataset were randomly selected to achieve similar number of slices with high-grade and low-grade gliomas. As batch normalization is a feature of “pix2pix”9, the effect of batch size on cGAN performance was studied. Training batch sizes of 1, 10, 100 and 200 were compared for each dataset. Given the large variation in image quality and contrast within datasets, the effects of image pre-processing through histogram equalization was explored. Histogram equalization was performed in Python 3 using the scikit-image package. For transfer learning, networks were pre-trained on BRATS-trained networks (batch size = 100, low-grade, high-grade and mixed, both original and histogram equalized) and network fine-tuning was performed on the meningioma training dataset (batch size = 1).
Testing: Dice-Sorensen coefficient (DSC) and Jaccard Index (JI) were used to evaluate segmentation accuracy.
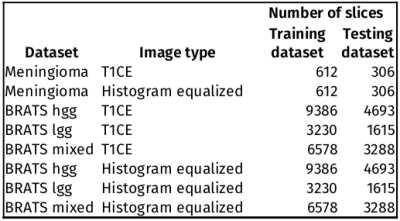
Table 1 shows the number of 2D slices in each training and testing dataset.
Results
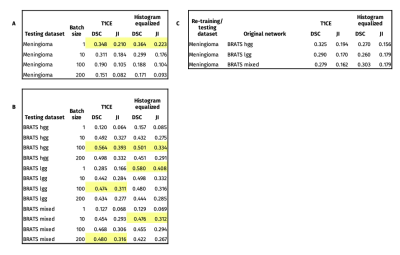
Table 2 shows DSC and JI training results. Figure 1 and 2 show selected segmentation images. Without histogram equalization, the highest DSC results were achieved for batch size = 1 (DSC = 0.347 ) in the meningioma dataset, batch size = 100 for high grade (DSC = 0.564) and low-grade (DSC = 0.474), and batch size = 200 for mixed (DSC = 0.468) BRATS datasets. Histogram equalization improved segmentation accuracy for meningioma (batch size = 1 and 200), low-grade BRATS (all batch sizes), high-grade BRATS (batch size = 1) and mixed BRATS (batch sizes = 1 and 10) training. The largest improvement with histogram equalization was seen for low-grade glioma (batch size = 1, from DSC = 0.285 to DSC = 0.579).Discussion
This work explored how different characteristics of a 2D cGAN training affect brain meningioma automated segmentation accuracy. We found that both batch size = 1 and batch size = 100 showed best results for meningioma and BRATS datasets testing, respectively. Histogram equalization improved learning in meningioma and low and mixed grade datasets. Network pre-training on glioma BRATS dataset did not improve meningioma segmentation results. This might be due to differences between the source images, segmentation objective and disease pathophysiology. We predict that further work to increase the number of training masks (local meningioma dataset) as well as image processing (skull stripping4) will allow better training and achieve higher segmentation accuracy. We expect improved results with the application of 3D cGANs, which have been shown to obtain higher DSCs4, but also would be more difficult to apply to routine MRI that includes 2D imaging.Conclusion
We found better brain meningioma MRI segmentation accuracy with 1) smaller batch sizes for smaller datasets and 2) image pre-processing with histogram equalization. Mixed trends in our results suggest that specific parameters of a network training should be tailored to the particular characteristics of the target dataset.Acknowledgements
This work was supported by the National Institute of Health Research Cambridge Biomedical Research Centre, Addenbrooke’s Charitable Trust and GlaxoSmithKline.References
1. Nowosielski M, Galldiks N, Iglseder S, et al. Diagnostic challenges in meningioma. Neuro Oncol 2017; 19: 1588–1598.
2. Kaus MR, Warfield SK, Nabavi A, et al. Segmentation of meningiomas and low grade gliomas in MRI. Lect Notes Comput Sci (including Subser Lect Notes Artif Intell Lect Notes Bioinformatics) 1999; 1679: 1–10.
3. Kaus MR, Warfield SK, Nabavi A, et al. Automated segmentation of MR images of brain tumors. Radiology 2001; 218: 586–591.
4. Laukamp KR, Thiele F, Shakirin G, et al. Fully automated detection and segmentation of meningiomas using deep learning on routine multiparametric MRI. Eur Radiol 2019; 29: 124–132.
5. Crimi A, Bakas S, Kuijf H, Menze B, Reyes M. Brainlesion: Glioma. InMultiple Sclerosis, Stroke and Traumatic Brain Injuries. In: Third International Workshop, BrainLes 2017.
6. Menze BH, Jakab A, Bauer S, et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans Med Imaging 2015; 34: 1993–2024.
7. Mirza M, Osindero S. Conditional generative adversarial nets. arXiv preprint arXiv:1411.1784. 2014 Nov 6.
8. Isola P, Zhu JY, Zhou T, et al. Image-to-image translation with conditional adversarial networks. In: Proceedings - 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017. Institute of Electrical and Electronics Engineers Inc., 2017, pp. 5967–5976.
9. Ioffe S, Szegedy C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. 32nd Int Conf Mach Learn ICML 2015 2015; 1: 448–456.
Figures