1887
Synthetic CMB generation for Training classifiers on QSM images1CSIRO Health and Biosecurity, Brisbane, Australia, 2Griffith University, Brisbane, Australia, 3Department of Nuclear Medicine and Centre for PET, Brisbane, Australia, 4University of Melbourne, Parkville, Australia, Melbourne, Australia, 5Austin Health Heidelberg, Australia, Melbourne, Australia, 6Griffith University, Gold coast, Australia, 7Data61, Brisbane, Australia
Synopsis
The lack of clinical dataset with enough examples of rare lesions challenges supervised machine learning methods. Here we propose to generate synthetic lesions for training a classifier to identify microbleeds from MRI QSM. We show that the performance of the classifier is improved compare to standard data augmentation using actual data alone. Our synthetic dataset can have unlimited size allowing to perform validation experiment, while keeping the actual data for testing. Moreover, many aspects of the data can be investigated, which would not be possible when using actual lesions: lesions can be synthetize on any location, size, shape, and intensity.
1. Introduction:
Cerebral micro-bleedings (CMB) are small chronic brain hemorrhages caused by structural abnormalities of the small vessels1. CMB can be found in individuals suffering from cognitive disorders, stroke, Alzheimer and might become more prevalent with aging. CMB are valuable biomarkers of cardiovascular diseases to assess cognitive impairment, but there are difficult and time consuming to detect even for experience radiologists, and automated CMB detection has attracted much interest2. MRI quantitative magnetic susceptibility mapping (QSM)3, a technique based on gradient-recalled echo (GRE) can show CMB with a positive contrast, as opposed to the more standard susceptibility weighted imaging acquisition where CMB show as hypointense semi-spherical lesions. Machine learning techniques are well suited to recognize CMB lesions, but obtaining large enough datasets with high quality ground truth is challenging because of the low prevalence of CMB. In addition, CMB mimics (mostly blood vessels) far outnumber lesions and often yields highly imbalance datasets skewing training of supervised machine learning methods. In this paper, synthetic data generation is proposed to overcome those challenges. Our proposed model does not depend on the availability of the ground truth, and an unlimited number of synthetic microbleeds (sCMB) can be generated covering all diversity of shape, location, and intensity features.2. Method:
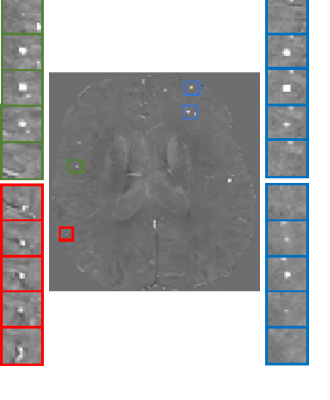
47 QSM and SWI scans were collected from 32 subjects, yielding 122 real microbleeds (rCMB) as visually assessed by an experience neurologist. rCMB are seen as a spherical-shaped and hypointense objects with small volume on SWI images. To generate sCMB on corresponding QSM images as hyperintense spherical-liked objects, we used a Gaussian shape in a high resolution patch [70x70x70], with random standard deviations ranging 0.7 to 0.9 inplane, while the standard deviation through slice was computed to obtain a given volume. Random shift (+/-5pixels) and random rotation (+/-30°) were added4. Partial volume effect was simulated by down sampling the patch with a factor of 10. The resulting mask was multiplied by actual patch randomly sampled from QSM of healthy individuals. In Fig.1, some samples of rCMB, generated sCMB and vessel cross section are presented. We used a simple feedforward artificial neural network (ANN) as a classifier and compared three models to address the imbalanced data problem: Model M1 under-sampled the majority class (negative) to have a balance training (61 positives and 61 negatives). Model M2 used the balance number of negatives and synthetic lesions (sCMB) as positives (2000 positives and 2000 negatives). Model M3 used traditional augmentation by flipping and rotating patches with rCMB with the same number of M2. The classifier was single-hidden layer feed-forward neural-network5, trained with scaled conjugate gradient. The number of epochs was 800, the mean square error was used as the cost function. The activation function in the hidden layer was Leaky Relu with 55 neurons. The training used 2-fold cross validation within 25 random draws (order of data). Ensemble learning was performed by averaging 15 networks for every draws.3. Result:
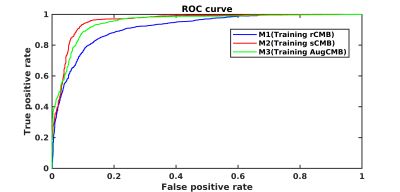
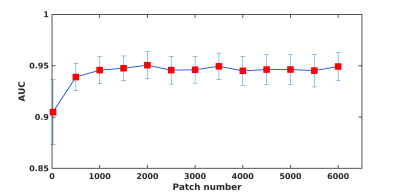
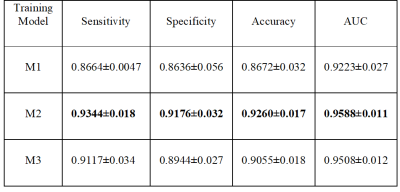
Results for the ANN classification on the testing datasets is reported in Table 1. Results shown are the mean and standard deviation over the draws for the area under the ROC (AUC), the sensitivity, the specificity, and the accuracy. Fig.2 shows the mean ROC for different training models. Using synthetic data outperformed traditional data augmentation. Fig.3 shows the AUC vs the number of sCMB training patches used in training in addition to the 60 rCMB patches. As can be seen, the use of additional sCMB patches in conjunction with the rCMB patches in training helps to improve the AUC from ~0.9 to ~0.95.4. Conclusion:
Lack of ground truth and small datasets for rare lesions in clinical application are common challenges for training AI system. In this paper, we propose the use of synthetic data as a data augmentation method for the automated classification of CMB from QSM. Advantages include unlimited dataset size, perfect ground truth, and large coverage of the feature space (e.g. wide range of volume, location, intensity…). In addition, all the training can be done using synthetic data, leaving the small dataset of actual lesions for testing. In our future work, we will explore using synthetic data for training more advanced classifiers based on deep learning.Acknowledgements
References
1. S. Martinez-Ramirez, S. M. Greenberg, and A. Viswanathan, “Cerebral mi-crobleeds: overview and implications in cognitive impairment,” Alzheimers Res. Ther., vol. 6, no. 3, p. 33, 2014.
2. Yates P, Sirisriro R, Villemagne V, Farquharson S, Masters C, Rowe C, et al. Cere-bral microhemorrhage and brain ˇ-amyloid in aging and Alzheimer disease.Neurology 2011;77(1):48–54.
3. Scott Ayton, Amir Fazlollahi, Pierrick Bourgeat, Parnesh Raniga, Amanda Ng, Yen Ying Lim, Ibrahima Diouf, Shawna Farquharson, Jurgen Fripp, David Ames, James Doecke, Patricia Desmond, Roger Ordidge, Colin L Masters, Christopher C Rowe, Paul Maruff, Victor L Villemagne, the Australian Imaging Biomarkers and Lifestyle (AIBL) Research Group, Olivier Salvado, Ashley I Bush, Cerebral quantitative susceptibility mapping predicts amyloid-β-related cognitive decline, Brain, Volume 140, Issue 8, Pages 2112–2119,August 2017.
4. S. Momeni, A. Fazllolahi, P. Bourgeat, P. Raniga, P. Yates, N. Yassi, P. Desmond, J. Fripp, Y. Gao,O. Salvado “Data augmentation using synthetic lesions improves machine learning detection of microbleeds from MRI ”,21 st International conference in medical image computing and computer assistant intervention (MICCAI), Granada Spain Sep 2018.
5. li Bin and R. Xuewen, “Review and performance analysis of single hidden layer sequential learning algorithms of feed-forward neural networks,” in 2013 25th Chinese Control and Decision Conference, CCDC 2013, 2013, pp. 2170–2175.
Figures