1886
Functional and structural connectivity predict MS patients’ impairment level using an ensemble model applied with a machine learning method1Department of Radiology, Weill Cornell Medicine, New York City, NY, United States, 2Judith Jaffe Multiple Sclerosis Center, Weill Cornell Medicine, New York City, NY, United States, 3Department of Neurology, Weill Cornell Medicine, New York City, NY, United States, 4Brain and Mind Research Institute, Weill Cornell Medicine, Ithaca, NY, United States
Synopsis
No study to date has performed a rigorous analysis of the relative contributions of multi-modal imaging data including the brain’s functional (FC) and structural connectivity (SC) in the task of classifying high and low adapting MS patients for a deeper understanding of the connectome-level mechanism contributing to variability in MS-related impairment. We built a machine learning based ensemble model that can accurately classify MS patients as high and low adapters (AUC> 0.626). We observed that SC and FC networks can be used to identify the most discriminative regions and to accurately classify MS patients regarding their impairment level.
Objective: We aim to build machine learning-based ensemble models that can accurately classify MS patients into two groups: high adapters (Expanded Disability Status Score (EDSS) < 2) and low-adapters (EDSS >= 2) using the FC and SC networks, identify the combination of imaging biomarkers that give the best accuracy in distinguishing MS patients by impairment severity, and to investigate which particular brain connections are most related to the disease in MS.
Material and Methods: Seventy-six MS patients (age: 45.37 ± 11.44 years, 66% female, disease duration: 12.48 ± 7.22 years) were included in our study; 23 had equal or higher than EDSS 2 at study baseline. The ensemble models were created by averaging the predictions of a machine learning method, Random Forest (RF), applied to three different simple models: (1) clinical model including age, sex, and disease duration, (2) FC model, and (3) SC model. SC and FC were measured between 86 Freesurfer based gray matter regions. Ensemble model 1 was created by averaging the predictions of all simple models, Ensemble model 2 was built by averaging the predictions of the clinical and SC models, and Ensemble model 3 was created by averaging the predictions of the clinical and FC models for each patient. Ensemble models averaged the predictions that were calculated as the probability of being low adapting for each MS patient. Then, the averaged probability was dichotomized with a threshold as high adapting ( < threshold) and low adapting ( >= threshold) MS patient. The performance of the ensemble models was assessed with the area under the ROC curve (AUC), balanced accuracy, sensitivity, and specificity.
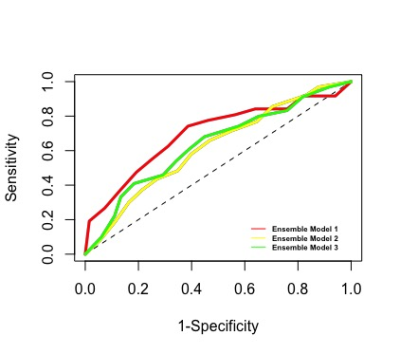
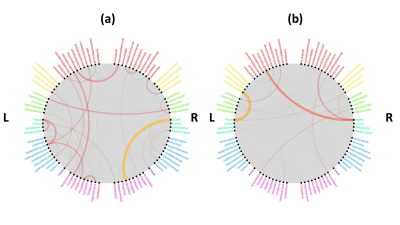
Results: Figure 1 shows that Ensemble model 1 that included the predictions of all simple models gave slightly higher AUC compared to two other ensemble models, however, there was no statistical difference between the AUC results of the ensemble models (Kruskal-Wallis test, p-value = 0.112). Ensemble model 1 that included clinical, SC, and FC performed well in classifying the high and low adapting MS patients with an AUC of 0.626 ± 0.093 and balanced accuracy of 0.632 ± 0.078. All ensemble models performed well in accurately classifying the low-adapting MS patients with a sensitivity higher than 0.726 ± 0.286. The most important clinical and demographic predictors were respectively age, disease duration, and spinal cord lesion number. Sex did not have high importance in predicting EDSS score. Figure 2 shows that the most discriminative SCs were between right lingual and ventral DC, between left lingual and superior temporal, and between left superior frontal and right rostral middle frontal. The most predictive FCs were found between left lingual and isthmus cingulate, between right lingual and left superior frontal, and between right lingual and superior frontal.
Conclusion: We observed that SC and FC networks can be used to identify the most discriminative regions and to accurately classify MS patients regarding their impairment level. The machine learning method performed in this study has the potential to help clinicians to predict the clinical impairment in MS patients, thus providing further information for personalized treatment decisions.
Acknowledgements
This work was supported by the NIH R21 NS104634-01 (A.K.) and NIH R01 NS102646-01A1 (A.K.).References
1. Barkhof, F. The Clinico-Radiological Paradox in Multiple Sclerosis Revisited. Current opinion in neurology 2002, 15, 239–45.
2. Schoonheim, M. M.; Meijer, K. A.; Geurts, J. J. G. Network Collapse and Cognitive Impairment in Multiple Sclerosis. Frontiers in neurology 2015, 6, 82.
3. Wojtowicz, M.; Mazerolle, E. L.; Bhan, V.; Fisk, J. D. Altered Functional Connectivity and Performance Variability in Relapsing-Remitting Multiple Sclerosis. Multiple Sclerosis Journal 2014, 20, 1453–1463.
4. Cruz-Gomez, A. J.; Ventura-Campos, N.; Belenguer, A.; Avila, C.; Forn, C. The Link between Resting-State Functional Connectivity and Cognition in MS Patients. Multiple Sclerosis Journal 2014, 20, 338–348.
5. Schoonheim, M. M.; Geurts, J. J. G.; Barkhof, F. The Limits of Functional Reorganization in Multiple Sclerosis. Neurology 2010, 74, 1246–1247.
6. Kuceyeski, A.; Shah, S.; Dyke, J. P.; Bickel, S.; Abdelnour, F.; Schiff, N. D.; Voss, H. U.; Raj, A. The Application of a Mathematical Model Linking Structural and Functional Connectomes in Severe Brain Injury. NeuroImage: Clinical 2016, 11, 635–647.
7. Rocca, M. A.; Valsasina, P.; Meani, A.; Falini, A.; Comi, G.; Filippi, M. Impaired Functional Integration in Multiple Sclerosis: A Graph Theory Study. Brain Structure and Function 2016, 221, 115–131.
8. Zurita, M., Montalba, C., Labbé, T., Cruz, J. P., da Rocha, J. D., Tejos, C., et al. (2018). Characterization of relapsing-remitting multiple sclerosis patients using support vector machine classifications of functional and diffusion mri data. Neuroimage Clin. 20, 724–730. doi: 10.1016/j.nicl.2018. 09.002
9. Kocevar, G., Stamile, C., Hannoun, S., Cotton, F., Vukusic, S., Durand-Dubief, F., et al. (2016). Graph theory-based brain connectivity for automatic classification of multiple sclerosis clinical courses. Front. Neurosci. 10:478.
10. Zhao Y, Healy BC, Rotstein D, Guttmann CR, Bakshi R, Weiner HL, et al. Exploration of machine learning techniques in predicting multiple sclerosis disease course. PLoS One. 2017;12(4):e0174866.
Figures