1877
Deep Learning Enables Accurate Quantitative Volumetric Brain MRI with 2x Faster Scan Times1Subtle Medical Inc, Menlo Park, CA, United States, 2RadNet, Encino, CA, United States, 3CorTechs Labs Inc, San Diego, CA, United States, 4Stanford University, Stanford, CA, United States
Synopsis
3D T1-weighted MRI is valuable for providing high resolution structural information and is commonly used in brain MRI exams despite long scan times. The recent development of deep learning (DL) has shown great potential for scan time reduction. However, the generalizability of DL methods is of concern for actual clinical deployment. In this study, we apply FDA-cleared DL software to accelerate 3D T1-weighted MRI scans by two folds and evaluate the quantification accuracy using FDA-cleared image analysis software. This study provides insight into the generalizability and accuracy of DL in clinical settings.
Purpose
3D T1-weighted MRI is valuable for providing high resolution structural information and is commonly used in neuro MRI exams despite long scan times. The availability of commercially available image processing software makes it possible to provide fast and accurate brain image analyses (e.g., image segmentation) and use them as biomarkers for various clinical indications [1-3]. The recent development of deep learning (DL) has shown great potential for image acceleration and image analysis [4]. However, the generalizability of DL methods is of concern for actual clinical deployment. In this study, we apply FDA-cleared DL software to accelerate 3D T1-weighted MRI scans by two times and evaluate the quantification accuracy using FDA-cleared image analysis software. This study provides insight into the generalizability and accuracy of DL in clinical settings.Methods
With IRB approval and patient consent, 32 subjects (age: 68$$$\pm$$$18 years; 19 male) undergoing clinical MRI exams were recruited. The study cohort includes cognitively normal, mild cognitive impairment, and Alzheimer’s disease subjects. Two T1-weighted volumetric scans were acquired for each subject: one from routine clinical protocol and the other with half the phase encodes and scan time. The faster scans were enhanced post acquisition by FDA-cleared DL software SubtleMR (Subtle Medical Inc, Menlo Park, CA). The paired datasets were collected on one of five different 3T scanners (3 GE and 2 Siemens). Both the standard images and DL-processed images were processed by FDA-cleared software NeuroQuant (CorTechs Labs Inc, San Diego, CA) for quantitative analysis. Results from the age related atrophy reports were compared. Hippocampal occupancy score (HOC), a biomarker to predict the progression of neurodegenerative diseases, as well as the volumes of hippocampi, superior lateral ventricles (SLV), and inferior lateral ventricles (ILV), were analyzed using linear regression, two-sided paired $$$t$$$-test and Bland-Altman analysis.Results
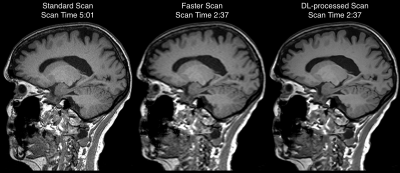
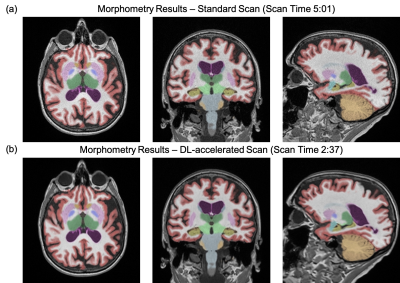
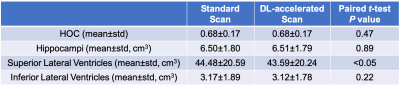
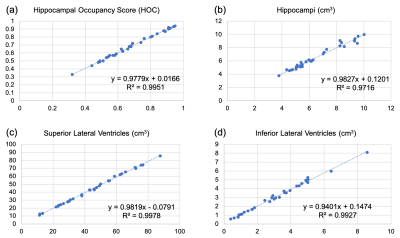
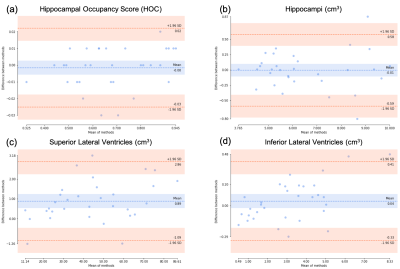
Example images from the standard scan, accelerated scan, and DL-processed scan are shown in Fig. 1. Excellent image quality (high SNR and image resolution) was obtained by DL. Example results of brain segmentation of the standard scan and DL-accelerated scan are shown in Fig. 2. Matching segmentation can be visualized. As shown in Fig. 3, the average HOCs did not differ: 0.68$$$\pm$$$0.17 and 0.68$$$\pm$$$0.17 for the standard scan and DL-accelerated scan, respectively. Paired $$$t$$$-tests also suggested that there was no statistical difference of HOC, hippocampal volume, and ILV (Fig. 3). The difference of SLV between two methods is approximately 2%. The scatter plots in Fig. 4 demonstrate strong correlation and linearity between two measurements. The Bland-Altman plots in Fig. 5 further demonstrate the strong agreement between two measurements.Discussion
This study has validated the high quantification accuracy of DL-accelerated scans with 2x faster scan times when compared with the standard longer scan. The range of HOC and strong agreement between standard scan and DL-processed scan has demonstrated the acceleration capability of DL in various neurogenerative disease conditions. Consistent results from scans on diverse scanner types demonstrated the good generalizability of the DL software. The inherent higher SNR from the DL processing could potentially improve the robustness of brain segmentation and will be the subject of future investigation.Conclusion
Deep learning can enable accurate quantitative volumetric brain MRI with 2x faster scan times.Acknowledgements
We would like to acknowledge the grant support of NIH R44EB027560.References
[1] McEvoy L, Brewer J (2012). Biomarkers for the clinical evaluation of the cognitively impaired elderly: amyloid is not enough. Imag. Med. 4:343-357.
[2] Heister D, et al. (2011). Predicting MCI Outcome with Clinically Available MRI and CSF Biomarkers. Neurology 77:1619-1628.
[3] Villemagne V, et al. (2013). Longitudinal assessment of neuroimaging and clinical markers in autosomal dominant Alzheimer’s disease: a prospective cohort study. Lancet Neurol. 12:357-367.
[4] Zaharchuk G, et al. (2018). Deep learning in neuroradiology. AJNR AM J Neuroradiol. 39:1776-1784.
Figures