1871
Realistic MRI simulation pipeline for anatomically variable normal young, aging and diseased brain1Biomedical Engineering, Eindhoven University of Technology, Eindhoven, Netherlands, 2MR R&D – Clinical Science, Philips Healthcare, Best, Netherlands, 3MR R&D – Collaboration Office, Philips Healthcare, Best, Netherlands, 4Philips Research Laboratories, Hamburg, Germany
Synopsis
A pipeline based on the XCAT phantom, the JEMRIS software for simulating the MR signal and a commercial reconstruction pipeline has been set-up for simulating realistic brain MRI images. Using this pipeline, an anatomically variable brain MRI population is simulated across age and gender. Anatomical variation is generated by means of changing individual brain sizes, and cortical gray matter volumes to mimic aging brain. MS lesions are simulated to mimic diseased brain as well. Significant contrast is generated across detailed brain structures. The commercial reconstruction pipeline increased the realism of simulated data.
Introduction
To make simulated MRI more realistic, a number of factors need to take into account. Including the use of finely structured phantoms with comprehensive tissue classes, true tissue properties, realistic simulation and realistic reconstruction. All these elements are integrated in our simulation pipeline by making use of XCAT phantom1, JEMRIS simulation software2 and clinically used Philips MRI reconstruction pipeline. The images generated using this pipeline, came out to be more realistic. Furthermore, to fill the gap of not having large sets of MRI data to be used for training and validating medical imaging analysis algorithms, a first set of anatomically variable simulated brain MRI images was created across age and gender using this pipeline. In previous studies3,4, only a limited number of brain sub-structures were simulated and no steps were taken to simulate full head and aging brain MRI. A greater number of simulated tissue classes and anatomical variability is required to test and optimize algorithms to segment respective tissues. Our simulations included comprehensive set of tissues, anatomical variability as well as pathology.Methods
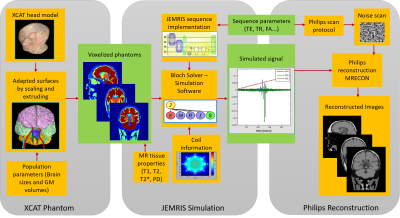
The pipeline designed for simulating realistic brain MRI is shown in Figure 1. The whole body 4D XCAT phantom1 for multimodality imaging research is used as starting point. The phantom includes highly detailed comprehensive male and female anatomies modeled as NURBS surfaces. The complete head model is shown in Figure 2.One type of anatomical variation is generated by scaling brain surfaces along anterior posterior, superior inferior and medial lateral directions. Five sets of head phantoms are generated for different brain sizes. Three of the selected scalings relate to the real measurements of subject’s brains from a 3D brain printing study5. The other two come from the default XCAT male and female anatomies, which are modelled according to the 50% percentile of US population. Brain measurements utilized across phantoms are presented in Figure 3b. To mimic aging brains, brain cortical Gray Matter (GM) volumes are adapted. Ten sets of head phantoms (five male and five female) with different corresponding GM volumes are generated by extruding and scaling each GM surfaces. Volumetric changes for normalized GM are taken from a study of tissue volumetric changes in young and aging population across gender6. Normalized GM measurements utilized across phantoms are presented in Figure 4. Furthermore, a head phantom with three spherical lesions of 3-5mm diameter, present in periventricular white matter is generated.
All generated 16 phantoms are voxelized at an isotropic resolution of 0.5mm3 from the surfaces. For a proof of principle, one slice per phantom in axial, sagittal and coronal view is selected for simulation. Diverse tissue properties from literature7,8 are assigned to the voxelized phantoms. 2D gradient echo T1w MRI images are simulated using open-source numerical Bloch-solver simulation software, JEMRIS2. Sequence parameters used are TE 10ms, TR 400ms and FA 90o, Sinc RF pulse of 2kHz bandwidth, max gradient strength of 22mT/m, max gradient slew rate of 100T/m/s and a uniform transmit and receive coil is used for simulation sequence design. Cartesian k-space data is simulated at 1mm2 resolution from a high resolution phantom model. To include realistic reconstruction, the raw k-space data is fed into the reconstruction pipeline that is used on Philips clinical MRI scanners. Simulated images are qualitatively evaluated for the generated contrast of brain details present, and the reconstruction image quality. To validate GM volumetric changes, cortical thickness is measured across simulated images.
Results
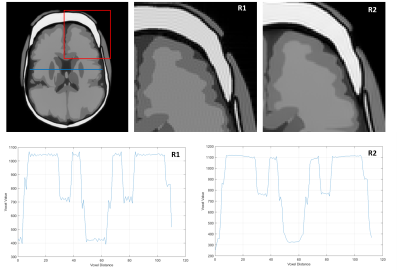
Simulated MRI samples for two (P1-2) full head phantoms with different brain sizes are presented in Figure 3a. In addition to the complete head structures and brain soft tissues, deep gray structures like putamen, thalamus, globus pallidus and caudate nucleus are visible in the slice due to significant contrast generated. In Figure 3c, simulated MRI sample with Multiple Sclerosis lesion (P3) is presented with all lesions visible. Another two (P4-5) simulated MRI samples for different GM volumes are presented in Figure 4. A cortical thickness decrease of 0.5mm in superior frontal gyrus is measured. Per slice simulation took ~20 min on 16 core processor. Reconstruction comparisons of simple FFT (R1) and Philips reconstruction pipeline (R2) for simulated k-space data are presented in Figure 5. Significant reconstruction improvement in R2 is visible as smooth boundaries and reduced Gibbs artifacts.Discussion and Conclusion
Using this pipeline, all anatomical variations are realistically represented in the simulated images. Detailed brain phantom and true relaxation times for each deep gray structure, has provided a significant contrast for deep gray structures visualization. Using clinically used realistic reconstruction pipeline, has generated simulated images with reduced artifacts, and fine contrasted edges with limited partial volume effects. They contain no variations yet within tissues due to lacking texture information. In addition, no noise and field inhomogeneities were yet been incorporated into our simulations.In the future, tissue texture information, partial volume and realistic noise need to be incorporated into our pipeline to make simulated image appearance even more realistic. The population of phantom instances has to be enlarged to represent further variability in terms of reflecting normal brain anatomical variations and brain pathologies. A more detailed aging brain with other structural changes corresponding to cortical GM volumetric changes has to be simulated.
Acknowledgements
This work has been supported by openGTN (opengtn.eu) project grant in the EU Marie Curie ITN-EID program (project 764465).References
1. Segars, W. P., et al. "4D XCAT phantom for multimodality imaging research." Medical physics 37.9 (2010): 4902-4915.
2. Stöcker, T., et al. "High‐performance computing MRI simulations." Magnetic resonance in medicine 64.1 (2010): 186-193.
3. Cocosco, C. A., et al. "Brainweb: Online interface to a 3D MRI simulated brain database." NeuroImage. 1997.
4. Alfano, B., et al. "An MRI digital brain phantom for validation of segmentation methods." Medical image analysis 15.3 (2011): 329-339.
5. Naftulin, J. S., et al. "Streamlined, inexpensive 3D printing of the brain and skull." PloS one 10.8 (2015): e0136198.
6. Farokhian, F., et al. "Age-related gray and white matter changes in normal adult brains." Aging and disease 8.6 (2017): 899.
7. Bojorquez, J. Z., et al. "What are normal relaxation times of tissues at 3T?." Magnetic resonance imaging 35 (2017): 69-80.
8. Okubo, G., et al. "Relationship between aging and T1 relaxation time in deep gray matter: A voxel‐based analysis." Journal of Magnetic Resonance Imaging 46.3 (2017): 724-731.
Figures