1870
Fetal Cortical Plate Segmentation using 2D Recurrent Residual U-Net with Plane Aggregation1Division of Newborn Medicine, Boston Children's Hospital, BOSTON, MA, United States, 2Hanyang University, Seoul, Korea, Republic of
Synopsis
Quantitative measurements about sulcal patterns are a basic approach to characterize the possibility of neurological disorders. In this study, we used recurrent residual U-Net. Also, to improve the accuracy of segmentation, the models of each plain in the MR image were produced separately and the results combined. As a result, the cortical plate segmentation showed an average dice coefficient of 0.901 ± 0.030, and segmentation for the internal region of the cortical plate showed 0.978 ± 0.010. The proposed method is expected to be useful as a quantitative developmental measurement of the fetus.
Introduction
Brain segmentation, including brain extraction, tissue segmentation, or region-of-interest (ROI) segmentation is fundamental for various analyses in magnetic resonance imaging (MRI) data. With advances in MRI technology, fetal brains can be visualized and observed like an adult brain, thereby MRI studies of brain development during the fetal period have been of great interest in recent. Quantitative measurements of cortical sulcal folding shape and pattern are important for characterizing normal and abnormal brain development and the possibility of neurological disorders.1-4 Previous studies have attempted to segment the cortical plate in the fetus by using the algorithms that have been used for adult MRI segmentation, or semi-automatic methods. Recently, deep learning has attracted significant attention in automatic medical image segmentation, but there is still a lack of its application for fetal brain image. In this study, we used the recurrent residual U-Net model proposed in R2U-Net for automatic cortical plate segmentation.5,6 In addition, to improve the accuracy of segmentation, segmentation on each axial, sagittal, and coronal plains were performed separately and the three segmentation results were combined.Method
2.1 DatasetThe dataset consists of 52 T2 MRI scanned on a 3T scanner at Boston Children’s Hospital (age: 26.7±2.5 gestational week [GW]). We performed manual segmentation for the cortical plate and inner region of the cortical plate. For the learning and performance verification, 10-fold cross-validation was performed considering GW. Flipped images were added to increase the amount of data for deep learning.
2.2 Preprocessing
Preprocessing steps for fetal brain MRI are described in 7. First, motion artifact on raw fetal brain MR images was corrected by combining multiple 2D slices.8 Then, we manually aligned the volume images along the anterior and posterior commissure. The registration to the standard template was performed to adjust the size between images and to align the position of the brain. Finally, to match the intensity distribution of the image between subjects, the intensity value was converted to the z-score for normalization.
2.3 Architectural design
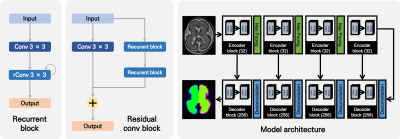
The proposed network has an encoder/decoder like 2D U-Net CNN architecture with 4 encoders and 4 decoders separated by the max-pooling/deconvolution layer (Figure 1). One block consists of one convolution followed by two recurrent convolution block and residual connection from input. The kernel was initialized in He normal method, and after the convolution, the activation function was applied elu and batch normalization.9-11 The network includes skipping connections between all encoder and decoder blocks of the same block depth, conceptually similar to basic U-Net architecture. The 2D input MRI slices and their corresponding segmentation maps are used to train the network with Adam optimizer implementation of Keras.12 The size of the batch was 30, and learning was performed using TITAN V GPU.
2.4 Loss function
In the medical imaging field, loss function for segmentation usually uses the dice coefficient. However, the dice coefficient is unfavorable to small structures as misclassifying a few pixels can lead to a large decrease in the coefficient. Therefore, to overcome the drawback of the dice coefficient, we use the logarithmic dice coefficient which more focus on incorrectly estimated labels.13 The loss defined as equation
$$L = \frac{\sum_{i}^R{(-\ln(Dice_{i} )^{0.3}}}{R}$$
In this paper, the cortical plate has a higher gradient because they are smaller than the whole inside region of the cortical plate.
2.5 Output aggregation
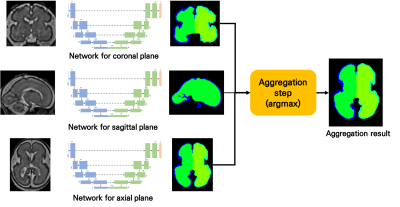
Although brain medical images consist of 3D, this paper proposed a 2D network. To apply a human-like approach to deep learning, this paper proposes a way to organize networks for each plane separately and combine the results made in each network. The result on the flip data were also used as a result of aggregation to improve the final results (Figure 2).
Results and Discussion
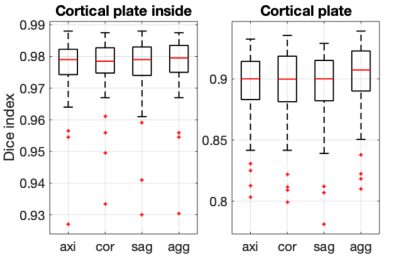
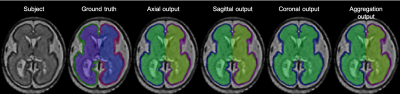
Our cortical plate segmentation showed an average dice coefficient of 0.901 ± 0.015 and segmentation for the internal region of cortical plates showed 0.978 ± 0.005. Output aggregation results showed significantly higher accuracies than axial, sagittal and coronal single plane results (Figure 3). The high accuracy in output aggregation can be considered as a refinement of the small mistakes that were made in the segmentation result from the one plane. The time required to learn is three times longer than to learn one plane. However, deep learning is not a big problem for practical use because it requires only a short time to produce the result after learning. The sample results are shown in (Figure 4).Conclusion
We have introduced 2D recurrent residual U-Net with a plane aggregation model for fetal cortical plate segmentation. The proposed network runs in seconds and shows good results. The proposed method is expected to be useful for the quantitative measurement of the fetal cortical plate. In the future, we will learn to reduce errors and provide more diverse segmentation results than we do now by adding surface distance loss functions that can minimize boundary distance errors between ground truth and prediction results.Acknowledgements
"This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number : HI19C0755)."
The Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health (NIH) (R21HD094130)
References
1. Im, K., Guimaraes, A., Kim, Y., Cottrill, E., Gagoski, B., Rollins, C., ... & Grant, P. E. (2017). Quantitative folding pattern analysis of early primary sulci in human fetuses with brain abnormalities. American Journal of Neuroradiology, 38(7), 1449-1455.
2. Im, K., Pienaar, R., Paldino, M. J., Gaab, N., Galaburda, A. M., & Grant, P. E. (2012). Quantification and discrimination of abnormal sulcal patterns in polymicrogyria. Cerebral cortex, 23(12), 3007-3015.
3. Tarui, T., Madan, N., Farhat, N., Kitano, R., Ceren Tanritanir, A., Graham, G., ... & Iyer, V. (2017). Disorganized patterns of sulcal position in fetal brains with agenesis of corpus callosum. Cerebral Cortex, 28(9), 3192-3203.
4. Ortinau, C. M., Rollins, C. K., Gholipour, A., Yun, H. J., Marshall, M., Gagoski, B., ... & Newburger, J. W. (2018). Early-Emerging Sulcal Patterns Are Atypical in Fetuses with Congenital Heart Disease. Cerebral Cortex.
5. Alom, M. Z., Hasan, M., Yakopcic, C., Taha, T. M., & Asari, V. K. (2018). Recurrent residual convolutional neural network based on u-net (R2U-net) for medical image segmentation. arXiv preprint arXiv:1802.06955.
6. Ronneberger, O., Fischer, P., & Brox, T. (2015, October). U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention (pp. 234-241). Springer, Cham.
7. Yun, H. J., Chung, A. W., Vasung, L., Yang, E., Tarui, T., Rollins, C. K., ... & Im, K. (2019). Automatic labeling of cortical sulci for the human fetal brain based on spatio-temporal information of gyrification. NeuroImage, 188, 473-482.
8. Kuklisova-Murgasova, M., Quaghebeur, G., Rutherford, M. A., Hajnal, J. V., & Schnabel, J. A. (2012). Reconstruction of fetal brain MRI with intensity matching and complete outlier removal. Medical image analysis, 16(8), 1550-1564.
9. He, K., Zhang, X., Ren, S., & Sun, J. (2015). Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. In Proceedings of the IEEE international conference on computer vision (pp. 1026-1034).
10. Clevert, D. A., Unterthiner, T., & Hochreiter, S. (2015). Fast and accurate deep network learning by exponential linear units (elus). arXiv preprint arXiv:1511.07289.
11. Ioffe, S., & Szegedy, C. (2015). Batch normalization: Accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv:1502.03167.
12. Kingma, D. P., & Ba, J. (2014). Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980.
13. Wong, K. C., Moradi, M., Tang, H., & Syeda-Mahmood, T. (2018, September). 3d segmentation with exponential logarithmic loss for highly unbalanced object sizes. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 612-619). Springer, Cham.
Figures