1862
Automatic detection of Multiple Sclerosis cortical lesions based on 3D-FLAIR and MP2RAGE sequences1LTS5, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland, 2Radiology Department, Center for Biomedical Imaging, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 3Universitäre Psychiatrische Dienste and University of Bern, Bern, Switzerland, 4Siemens Healthcare AG Switzerland, Lausanne, Switzerland, 5Neurologic Clinic and Policlinic, Departments of Medicine, Clinical Research and Biomedical Engineering, University Hospital Basel and University of Basel, Basel, Switzerland, 6Translational Imaging in Neurology (ThINk) Basel, Department of Medicine and Biomedical Engineering, University Hospital Basel and University of Basel, Basel, Switzerland
Synopsis
Multiple Sclerosis cortical lesions are not readily visible in conventional MRI, but they are clinically highly relevant and have been recently included in the MS diagnostic criteria. However, advanced MRI sequences such as the MP2RAGE are needed in order to identify them visually. In this work, we propose an automatic method based on a convolutional neural network to automatically detect cortical lesions. In a cohort of 84 patients with FLAIR and MP2RAGE acquisitions our framework achieves a 77% cortical lesion detection rate with a 26% lesion-wise false positive rate.
Introduction
Multiple Sclerosis (MS) patients present lesions in both the white and gray matter of the central nervous system. The automated detection of cortical lesions (CLs) in the brain is a challenging task that, contrarily to white matter lesions (WMLs), has not been largely explored yet1,2,3. CLs are not readily visible4 in conventional magnetic resonance imaging (MRI) such as magnetization-prepared rapid gradient-echo (MP-RAGE) or FLuid Attenuated Inversion Recovery (FLAIR) at 3T. Advanced sequences such as Magnetization-Prepared 2 Rapid Acquisitions Gradient Echo (MP2RAGE)5 provide a better contrast to visually identify them5. The count and volume of CLs are highly clinically relevant and were recently included in the diagnostic criteria for MS6. In this work, we present a fully convolutional neural network (CNN) for the detection of MS brain WMLs and especially CLs, using FLAIR, and MP2RAGE contrasts. Differently from previous works1,2,3, we evaluate the reported detection framework on a dataset of unprecedented size using image contrasts that are today available on many clinical scanners.Materials and Methods
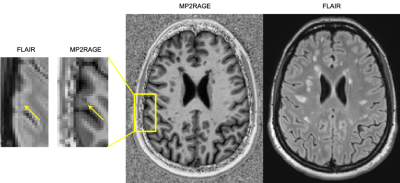
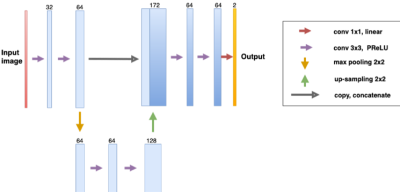
We include MRI data from 84 patients acquired in two hospitals. Dataset (1) consists of 36 patients from Lausanne University Hospital (20 female / 16 male, mean age 34±10 years, age range [20-60] years, mean EDSS 1.5±0.3) in the early stage of MS. Subjects were scanned at 3T (MAGNETOM Trio, Siemens Healthcare, Erlangen, Germany) using a 32-channel head coil. The protocol included 3D FLAIR (TR/TE/TI=5000/394/1800ms), and MP2RAGE (TR/TI1/TI2=5000/700/2500 ms) contrasts (resolution=1.0x1.0x1.2 mm3). Dataset (2) consists of 48 patients from University Hospital Basel (32 female / 16 male, mean age 47±13 years, age range [26-73] years, mean EDSS 3.5±2.0). Subjects were scanned at 3T (MAGNETOM Prisma Siemens Healthcare, Erlangen, Germany) using a 64-channel head coil. This protocol included FLAIR (TR/TE/TI=5000/386/1800 ms), and MP2RAGE (TR/TI1/TI2=5000/700/2500 ms) contrasts (resolution=1.0x1.0x1.0 mm3). Each dataset had experts’ manual reference annotations of MS lesions (WMLs and different types of cortical lesions) that we used as ground truth for training and testing. Overall the experts found a total of 3416 WMLs and 772 CLs (662 of type I and 110 of type II). An example of the two contrasts and the corresponding reference annotation of a CL is shown in Figure 1.Pre-processing steps included rigid registration of FLAIR images to MP2RAGE space using ELASTIX and N4 intensity normalization7. To automatically segment MS lesions, we propose a CNN based on the 3D U-Net architecture8. Differently from the original 3D U-Net, our architecture (Figure 2) had only two resolution levels in order to prevent overfitting. The window input size was (76,76,76) and the output (60,60,60). Moreover, we implemented a sampling scheme which allows a balanced training for all structures of interest, regardless of their size. Data augmentation such as rotations, and flipping was applied, again to prevent overfitting. The network was trained to minimize the cross-entropy loss with a batch size of 2 using Adam as optimizer. Training was performed mixing dataset (1) and (2), and we evaluated our approach in a 5-fold cross-validation fashion considering the detection rate (DR) per lesion type, lesion-wise false positive rate, Dice coefficient (DSC), and volume difference (VD). The code has been implemented in NiftyNet9 running on top of Tensorflow10.
Results
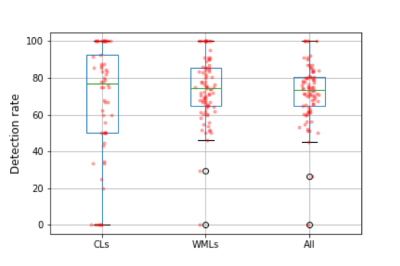
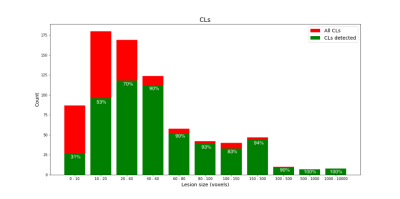
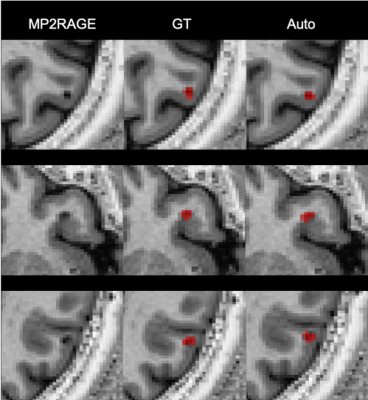
The proposed method achieves a median CL detection rate of 77%, a median WML detection rate of 74%, lesion-wise false positive rate of 26%, DSC of 54%, and VD of 43%. Boxplots of the detection rate for the different types of lesions are shown in Figure 3. The number of samples in the boxplot of CLs is lower since CLs were not identified in some of the patients in our cohort. As expected, detection rate of CLs is higher for bigger lesions (Figure 4). Figure 5 shows examples of manual and automated segmentation of CLs.Discussion and Conclusion
In this work, we explored the capability of deep-learning-based techniques for the automated detection of different types of MS lesions (WMLs and CLs) using conventional and advanced MRI sequences at 3T (FLAIR, MP2RAGE) of 84 MS patients. To the best of our knowledge, this is the largest cohort considered for automated detection of cortical lesions. Our method achieved a high detection rate for all types of MS lesions (73%), and for CLs (77%) with a relatively low lesion-wise false positive rate (26%). These results are similar to those obtained in previous studies1,2,3 aiming at CL segmentation. However, these studies considered smaller datasets and included additional non-standard imaging contrasts such as double inversion recovery (DIR). Our approach is based only on two sequences, which facilitates the translation into clinical practice. A limitation of our work are the results in terms of segmentation, since our CNN achieves a relatively low DSC of 54% and a VD of 43%. Future studies will therefore aim at improving the lesion delineation as well as focusing on achieving a higher detection rate of very small CLs.Acknowledgements
This project is supported by the European Union's Horizon 2020 research and innovation program under the Marie Sklodowska-Curie project TRABIT (agreement No 765148). The work is also supported by the Centre d'Imagerie BioMedicale (CIBM) of the University of Lausanne (UNIL), the Swiss Federal Institute of Technology Lausanne (EPFL), the University of Geneva (UniGe), the Centre Hospitalier Universitaire Vaudois (CHUV), the Hôpitaux Universitaires de Genève (HUG), and the Leenaards and Jeantet Foundations. CG is supported by the Swiss National Science Foundation grant SNSF Professorship PP00P3-176984. AA was supported by the Swiss National Science Foundation grant SNSF 173880.References
1. Fartaria, Mário João, et al. "Automated detection of white matter and cortical lesions in early stages of multiple sclerosis." Journal of Magnetic Resonance Imaging 43.6 (2016): 1445-1454.
2. Fartaria, Mário João, et al. "Segmentation of cortical and subcortical multiple sclerosis lesions based on constrained partial volume modeling." MICCAI. Springer, Cham, 2017.
3. La Rosa, Francesco, et al. "Deep learning-based detection of cortical lesions in multiple sclerosis patients with FLAIR, DIR, and MP2RAGE MRI sequences." Multiple Sclerosis Journal 25.CONF (2019): 131-356.
4. Calabrese, Massimiliano, Massimo Filippi, and Paolo Gallo. "Cortical lesions in multiple sclerosis." Nature Reviews Neurology 6.8 (2010): 438.
5. Kober, Tobias, et al. "MP2RAGE multiple sclerosis magnetic resonance imaging at 3 T." Investigative radiology 47.6 (2012): 346-352.
6. Thompson, Alan J., et al. "Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria." The Lancet Neurology 17.2 (2018): 162-173.
7. Tustison, Nicholas J., et al. "N4ITK: improved N3 bias correction." IEEE transactions on medical imaging 29.6 (2010): 1310.
8. Çiçek, Özgün, et al. "3D U-Net: learning dense volumetric segmentation from sparse annotation." International conference on medical image computing and computer-assisted intervention. Springer, Cham, 2016.
9. Gibson, Eli, et al. "NiftyNet: a deep-learning platform for medical imaging." Computer methods and programs in biomedicine 158 (2018): 113-122.
10. Abadi, Martín, et al. "Tensorflow: A system for large-scale machine learning." 12th {USENIX} Symposium on Operating Systems Design and Implementation ({OSDI} 16). 2016.
Figures