1798
180 vertebrate brains for an open data set of comparative neuroanatomy1Groupe de Neuroanatomie appliquée et théorique, Unité de Génétique humaine et fonctions cognitives, Département de neuroscience, Institut Pasteur, Paris, France, 2Centre de Neuroimagerie de Recherche, Institut Cerveau Moelle – ICM, CENIR, UPMC-Inserm U1127, CNRS 7225, Paris, France, 3Département Adaptations du Vivant, UMR MECADEV 7179 Equipe FUNEVOL Sorbonne Universités-MNHNUPMC- CNRS-IRD, Muséum National d'Histoire naturelle, PAris, France, 4Center for Research and Interdisciplinarity (CRI), Université Paris Descartes, Paris, France
Synopsis
This work presents the MR acquisition of 180 post mortem brains from different species coming from a Museum collection. 3D-High resolution images were obtained at 3 and 11.7T, depending on brain size. The images where then preprocessed in order to be available online for dowload on The Brain Catalogue (https://braincatalogue.org) web portal designed for comparative neuroanatomy studies.
Introduction
Most studies of brain evolution are limited by three main factors. One is that brain MRI data available for research mostly comes from primates or rodents. Another one is that when data from less common species is available, reported volumetric measurements are mostly based on extrapolations from unavailable 2D histological slices. Finally, data are usually not shared, which limits the emergence of new concepts or hypotheses in comparative neuroanatomy1 .Compared to other techniques, MRI allows us to obtain very detailed measurements of brain anatomy of old postmortem samples in a relatively easy fashion and reasonable time2. During the last 5 years we have successfully obtained high quality MRI of 180 brains of different species, coming from the collection of brains in fluid from the French National Natural History Museum (MNHN). We are working towards making that data available online in an open web portal: the Brain Catalogue (https://braincatalogue.org). A main objective of this work is to help reinvigorate research in comparative neuroanatomy, by feeding the Brain Catalogue – which offers tools for visualisation and manual segmentation of the data. This unique and high-quality data set should help considering an evolutionary and phylogenetic background in the study of brain variability.We describe some of the approaches that have allowed us to scan this large sample of brains from museum specimens.Materials and Methods
Due to the age of the collection (end of 19th – early 20th century) and the lack of information about the methods used to extract and preserve brains, brains were scanned within their original jars to prevent degradation or air contamination. Some of the brains in the collection are very rare, like that of the extinct Thylacine3. MRI was performed using a 3T Prisma system (Siemens, Germany) or an 11.7T Bruker Biospec (Bruker, Germany) using 3D gradient-echo sequences (FLASH). The choice of the sequence was motivated by its relative independence to the conservative liquid. Field of view (FOV) and matrix size were adjusted to the size of the specimen and were chosen to obtain the highest resolution achievable with our systems (from 25 µm3 for rodents at 11.7T to 450 μm3 for an elephant at 3T). TR and TE were always selected to the minimum possible value. The number of averages was chosen to maintain a scanning time below 12 hours (we scanned overnight). Depending on the size of the jar containing the specimen, different coils were used: At 3T, Body Coil was used for emission and Flex 4-channel coils or Special Purpose coils were used for signal reception. At 11.7T, a 72 or 38-mm birdcage resonator or mouse cryoprobe for both emission/reception were used. After scanning and visual quality check (QC), a series of preprocessing steps are being performed with in-house tools first to perform automated and unbiased QC , then to reorient the brains in a standard manner, image cropping and finally to provide semi-automatic segmentation of the brain tissue.Results
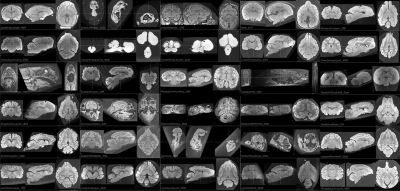
Brains of 180 different species were successfully scanned and passed QC. Images are currently being preprocessed and will be uploaded to the Brain Catalogue, where they will be available for download in Nifti format. Examples are shown in Figure 1.Discussion and Conclusion
We describe the MRI scanning procedure of a unique set of postmortem brains from 180 different species. This dataset will be progressively made available online in the Brain Catalogue. Our results show that high quality data can be obtained from old natural history collections, without any damage to the tissue. In cases were the brains can be taken from their containers, it would be possible to enhance the contrast and signal-to-noise using methods such as Gadolinium soaking4, for example. Comparative brain data will allow researchers to study a variety of questions concerning brain variability and brain evolution.Acknowledgements
This work was suported by the e-museum program. MNHN, France.References
1. Heuer et al., “Evolution of neocortical folding: A phylogenetic
comparative analysis of MRI from 34 primate species”, Cortex, Apr 2019. DOI: 10.1016/j.cortex.2019.04.011.
2. Santin et al., “The MRI of Darwin: The Brain Catalogue”, ISMRM 2014, Milano, Italy.
3. Santin et al., “Brain Catalogue and its MRI of extinct species: the example of Thylacinus Cynocephalus”, ISMRM 2016, Singapore.
4. Sébille et al., "“Post mortem high resolution tractography of the macaque brain with 11.7T MRI”, J. Neuroscience Method, Jan 2019. DOIi: 10.1016/j.jneumeth.2018.10.010.