1790
3D volume reconstruction from three orthogonal multi-slice 2D images using a super-resolution network
Xue Feng1, Huitong Pan2, Li Zhao3, and Craig H. Meyer1
1Biomedical Engineering, University of Virginia, Charlottesville, VA, United States, 2Springbok, Inc., Charlottesville, VA, United States, 3Children's National Health System, Washington DC, DC, United States
1Biomedical Engineering, University of Virginia, Charlottesville, VA, United States, 2Springbok, Inc., Charlottesville, VA, United States, 3Children's National Health System, Washington DC, DC, United States
Synopsis
High-resolution 3D MRI can provide detailed anatomical information and is favorable for accurate quantitative analysis. However, due to the limited data acquisition time and other physical constraints such as breath-holding, multi-slice 2D images are often acquired. The 2D images usually have a larger slice thickness than the in-plane resolution. To reconstruct the high- resolution 3D MRI, we propose to use a super-resolution network with three orthogonal multi-slice 2D images as the input. We validated the proposed method on brain MRIs and achieved good results in terms of mean absolute difference, mean squared difference and image details with visual inspection.
Introduction
3D magnetic resonance images (MRI) with high spatial resolution can provide detailed anatomical information and are favorable for accurate quantitative analysis. However, due to the limited data acquisition time and other physical constraints such as breath-holding, multi-slice 2D images are much easier to acquire. The 2D images usually have a larger slice thickness than the in-plane resolution. To reconstruct a high-resolution (HR) 3D MRI volume, various super-resolution image processing methods have been proposed, primarily based on linear interpolation. Recently deep learning methods have been proposed to up-sample 2D images using super resolution networks; however, most of these methods have only used one stack of 2D images as the input. As three orthogonal multi-slice 2D images are often acquired in practice, we hypothesize that using them as the input images can improve the fidelity of volume reconstruction. In this study, we aim to develop a super resolution network that can take the input from three orthogonal multi-slice 2D images and accurately reconstruct 3D volumes. Its performance in brain MRI will be validated.Methods
The data we used in this study included 107 isotropic T2 brain MRIs with 1x1x1 mm3 resolution from MIDAS [1]. 85 were randomly selected for training and the remaining 22 for validation. The original isotropic MRIs were regarded as the ground-truth high-resolution (HR) images. For training purposes, the original HR images were cropped into patches of 80x80x80. To simulate the multi-slice 2D images, we down-sampled the HR images along three orthogonal directions. For each direction, we averaged every 5 HR slices to form 1 slice. Averaging was used instead of resizing to simulate the actual 2D data acquisition process with sinc-based slice-select RF pulses. Therefore, for each HR patch, we obtained 3 multi-slice 2D patches with the dimensions of 16x80x80, 80x16x80, and 80x80x16, respectively. To reconstruct the HR patch, linear interpolation was first used to up-sample the three down-sampled images to the desired HR dimension. A 3D encoding-decoding network based on the U-Net structure was built with the 3 up-sampled images concatenated along the last dimension (80x80x80x3) as the network input. In addition, a generative adversarial network (GAN) was built with the 3D U-Net as the generator and a CNN as the discriminator.Results
The mean absolute difference (L1) and mean squared difference (L2) between the model outputs and the HR targets for 22 validating subjects were calculated. All of the reconstruction networks were trained with 1500 epochs. To evaluate the linear interpolation performance, the L1 and L2 differences were calculated between the averaged up-sampled images and the ground truth as well. Furthermore, the performance of the 3D U-Net with 3 orthogonal inputs was compared with only the axial input.As shown in Table 1, the 3D U-Net with 3 orthogonal inputs outperforms linear interpolation and the 3D U-Net with a single input. It also has slightly lower L1 and L2 values than the GAN model. Figure 1 further demonstrates that 3D U-Net with 3 LR inputs is able to reconstruct more HR image details than the model with a single input and linear interpolation. GAN has a similar output to the 3D U-Net with minor improvements in image details, as seen near the arrow in the sagittal images. However, GAN may yield unrealistic imaging artifacts as demonstrated at the white boxes in the coronal and sagittal images.
Discussion & Conclusion
Experiment results demonstrate that 3D U-Net can reconstruct a significant amount of HR detail with 3 simulated LR images. The proposed method of HR MRI reconstruction can loosen the physical constraints for obtaining HR MRIs and allow accurate quantitative analysis when only LR MRIs are available. However, our simulation of the LR images may not be representative for the orthogonal MRIs acquired in actual practices, as our simulation did not reflect possible MRI artifacts, such as breathing motion and patient’s movement between scans. For future studies, we will test our model on data with those artifacts and further evaluate its performance on actual data.Acknowledgements
No acknowledgement found.References
- Bullitt E, Zeng D, Gerig G, Aylward S, Joshi S, Smith JK, Lin W, Ewend MG. Vessel tortuosity and brain tumor malignancy: A blinded study. Academic Radiology; 2005. 12:1232-1240.
Figures
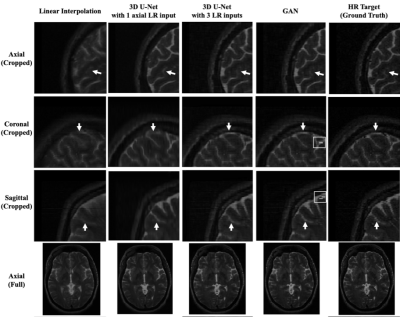
Figure 1. The top three rows are the cropped MRI patches
in the dimension of 80mm ´ 80mm ´80mm. The last row is the full axial MRI in the
dimension of 192mm ´ 256mm.
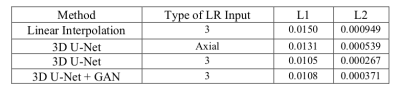
Table
1: Average validation loss. For linear interpolation with
3 LR inputs, the losses are the average losses of three interpolated outputs.