1733
Assessment of Cerebello-Thalamo-Cortical (CTC) Fiber Pathways in Post-Surgical Medulloblastoma Patients Using a CTC Template1Diagnostic Imaging, St.Jude Children's Research Hospital, Memphis, TN, United States, 2Psychology, St.Jude Children's Research Hospital, Memphis, TN, United States, 3Epidemiology and Cancer Control, St.Jude Children's Research Hospital, Memphis, TN, United States
Synopsis
A CTC fiber pathway template was built in MNI space from the CTC data of 10 healthy controls. The template was aligned with the principle diffusion directions of each individual DTI data of additional 10 healthy controls and 11 post-surgical medulloblastoma patients using a linear algorithm developed in this study. The aligned template can accurately mimic the real CTC pathways in an individual subject in both healthy and post-surgical subjects. The DTI parameter values in post-surgical medulloblastoma patients can be accessed using the transformed CTC pathway.
INTRODUCTION
CTC pathway is an important neural fiber tract that connects cerebellum with contralateral frontal cortex. The CTC damage due to surgery for mid-line intravascular tumor in posterior fossa can result in different degrees of tremor, ataxia and cerebellar cognitive affective syndromes (CCAS) such as seen in cerebellar mutism syndrome. A quantitative measure of evaluation for the CTC pathways in postoperative patients is necessary to understand the relationship between CTC damage and post-surgical CCAS. Diffusion tensor tractography (DTT) provides a possibility to identify the CTC, but can fail in native space if there is loss of diffusion information along the CTC path due to surgical damage. In this study, we proposed an alternative approach using a CTC pathway template from healthy subjects to access the damaged CTC in patients.METHOD AND MATERIALS
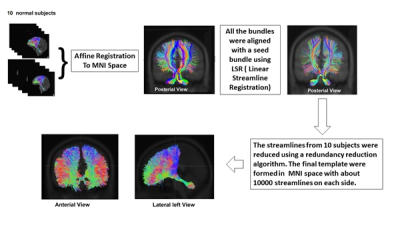
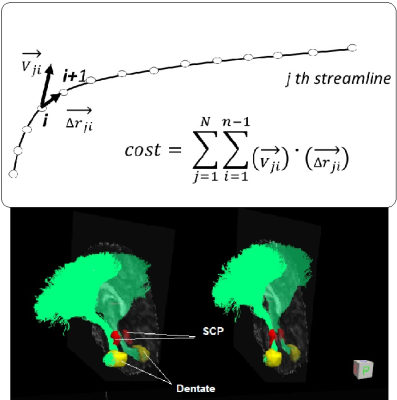
MR scans of 11 childhood medulloblastoma patients (age at exam 12.2±3.2 years) and 20 healthy controls (age 21±5.7 years) were used. Each MR scan consisted of an anatomic 3D T1 weighted image data set and a DTI data set (64 directions, 2 average, b=1500). For each subject, the left and right CTC pathways were extracted using the probabilistic fiber tracking scheme described in our previous study1. The CTCs of 10 healthy subjects were then used to build a CTC template. As shown in Figure 1, the CTC streamlines from 10 healthy subjects were first affine registered to MNI space. One registered CTC was chosen as a target template, and the rest of the CTCs were aligned with the target template using a bundle to bundle registration algorithm 2 which was implemented in DIPY (http://dipy.org). The final CTC template consists of 20,000 streamlines (10,000 on each side) with each subject contributing at least 10% to the final template. To align the template with an individual subject, we proposed a method that aligned the streamlines in the template with the native principle diffusion directions. The native principle diffusion direction V1 and fractional anisotropy (FA) images were first obtained using the DTIFit software in FMRIB Toolbox (http://fsl.fmrib.ox.ac.uk ). Affine registration of the MNI template with the native FA was then performed to transform the template to native DTI space. At this stage, the template was not always aligned well in the native space. A cost function to align the streamlines in the template with the native principle diffusion directions (V1) were proposed as illustrated in Figure 2. The template can be aligned in the native space by maximizing this cost function through rotation and translation of the template in native space. To validate this method, the CTC data of 10 additional healthy subjects and 11 post-surgical medulloblastoma subjects in native space were used.RESULTS
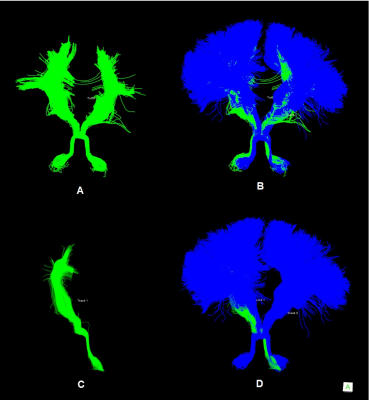
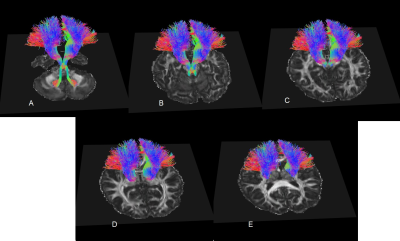
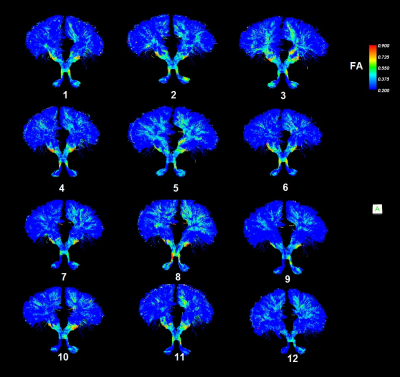
The bilateral CTCs of 20 heathy subjects were successfully obtained in native space. Expectedly, among 11 patient subjects, the bilateral CTCs could be tracked in only 5 subjects, left CTC could be tracked in an additional 4 subjects and neither left nor right CTCs could be tracked in 2 subjects. The CTC template was transformed into the native space for each subject. The bottom part of Figure 2 demonstrates the effect of aligning the streamlines of the template with the native principle diffusion directions by maximizing the cost function in native space. The aligned template on each subject was compared with the tracts natively obtained. Figure 3 demonstrates visual comparison of aligned templates with natively tracked CTCs of a healthy subject and a patient. The streamlines in the locally tracked CTC fiber bundle were not exactly the same as the streamlines in the aligned template, but provided a good approximation of the locally tracked bundles. Figure 4 illustrates the streamlines of the aligned template in a patient subject. It connected with dentate nuclei, superior cerebellar peduncles, contralateral red nucleus, thalamus and internal cuspules. With the template aligned in native space, the quantitative evaluation of the CTC can be performed on the aligned template using locally acquired DTI parameters. Figure 5 shows the FA distributions along the CTC pathways of 12 subjects (1 normal and 11 patients). For those 11 patient subjects, the CTC damage primarily occurred on the right CTC tracts. Since FA is an important parameter for the neural fiber integrity, the degree of the CTC damage can be quantitative evaluated on the FA statistics of the CTC tracts. An advantage of using one template for all subjects is the possibility of transforming all data into a common space for group analysis with tract-based morphometry.3DISCUSSION / CONCLUSION
A DTI model is insufficient to track CTCs when the seed region or tract itself is damaged by surgery. By applying a template, the loss of diffusion information along the tract in a post-surgical patient subject can be overcome. This study showed that the proposed method was feasible and the template could be successfully transformed into the native spaces of 21 subjects.Acknowledgements
No acknowledgement found.References
1. Ji Q., et al, “Measurement of projections between dentate nucleus and contralateral frontal cortex in human brain via diffusion tensor tractography“ Cerebellum. 2019, 18(4):761-769.
2. Garyfallidis E., et al “Robust and Efficient linear registration of white-matter fascicles in the space of streamlines” NeuroImage, 2015, 117:124-140
3. Lauren J. et al “Tract-based morphometry for white matter Group analysis”. NeuroImage, 2009, 45:124-140
Figures