1731
Texture-based radiomics of peritumoral tissue predict subsequent progression into cancer in Glioblastoma Multiforme1Radiology, Stony Brook University, Nesconset, NY, United States, 2Radiology, Stony Brook University, Stony Brook, NY, United States
Synopsis
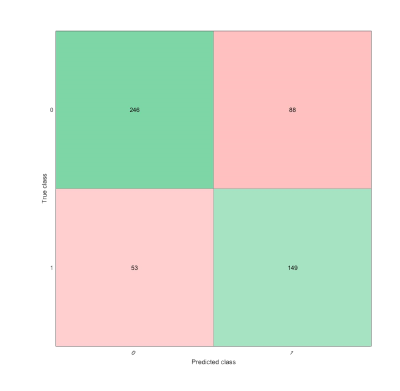
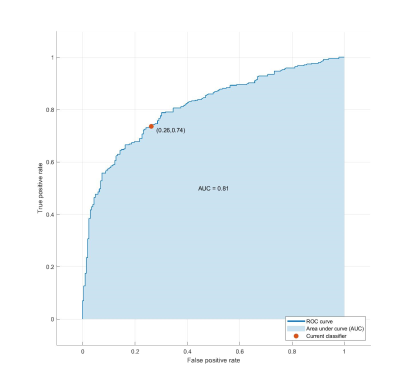
Tumor recurrence in Glioblastoma Multiforme patients is nearly inevitable. This study analyzes the texture-based radiomic features of the peritumoral tumor habitat to identify the distinguishing features of tissue conducive to tumor growth. Peritumoral region and FLAIR edema are segmented and divided into subregions, which are labeled positive if it contains many voxels that appear normal, but subsequently becomes tumor. Texture features are extracted from each subregion and selected for use in an SVM. The classifier achieved 74.81% accuracy and .81 AUC. Texture analysis of peritumoral region can predict cancer growth and enables better treatment and surgical planning.
Introduction
Tumor recurrence is nearly inevitable in patients diagnosed and treated for Glioblastoma Multiforme (GBM) because of the disorderly proliferation beyond surgical margins1. Current standard of care is maximum justifiable and safe resection of the enhanced tumor2. GBM, however, tends to expand into the peritumoral region over time, thus the peritumoral tumor habitat may have a distinct microenvironment conducive to subsequent tumor growth3. The aim of this study was to identify the texture features of the peritumoral region associated with subsequent progression to tumor.Methods
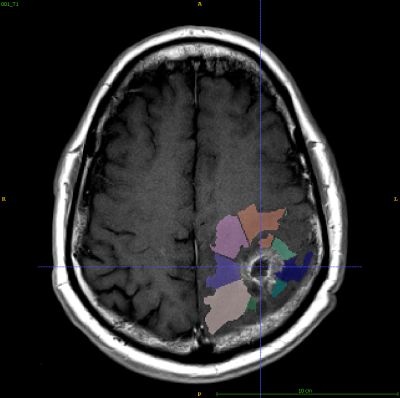
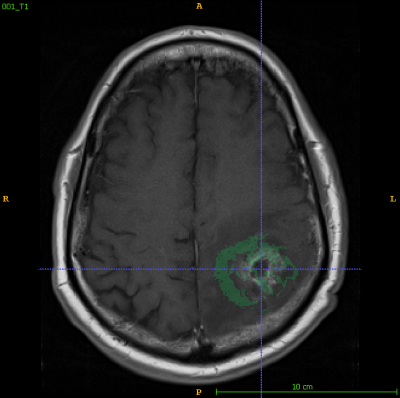
17 patient de-identified files with T1-weighted, FLAIR, and T2-weighted sequences from two timepoints (within 90 days post chemoradiotherapy completion and at progression) were obtained from The Cancer Imaging Archive (TCIA)4. All images coregistered to the first time point (Tp1) post-contrast T1-weighted image using FLIRT (FMRIB’s Linear Image Registration Tool)5. Tumors at both time points were segmented by experienced radiologists based on abnormal MRI characteristics and served as ground truth. Peritumoral regions (within 8mm of tumor edge) and FLAIR edemas were segmented in ITK-SNAP6 and divided into 16 patches using MATLAB7. Texture features(n = 75) were extracted for all patches larger than 64 voxels (n = 536) from their respective T1-ce images using PyRadiomics8. Each patch was labeled 1 if they contained more than 64 voxels that would progress into tumor and 0 if not. A subset of optimal features was selected LASSO regularization9 and used to train a Linear Support Vector Machine10 in MATLAB.Results
Of the 75 texture features, 11 features were selected by LASSO regularization. The linear SVM radiomics model was evaluated using leave-one-out cross validation and attained a 74.81% accuracy and an area under receiver operating characteristic curve (AUC or AUROC) of .81 for prediction of tissue that will become tumor on the validation set and an accuracy of 73.2% on separate augmented testing set.Discussion
The linear SVM radiomics model suggests that tissue susceptible of tumor growth have a distinct radiomic signature compared to low-risk tissue. The SVM also falsely predicts subsequent tumor regions at a higher rate than low-risk tissue. The limitations include a small sample size and low resolution of MR images.Conclusion
Texture analysis of peritumoral based on MRI tissue accurately predicts cancer progression. Knowledge of tissue that will likely become tumor based on non-invasive MRI enables better prognosis and treatment planning. Further investigation would implement more sophisticated texture features and segmentation algorithms to find tissue likely to become tumor with greater detail.Acknowledgements
I would like to express my gratitude towards Dr. Haifang Li and Mr. Zenzerovich for their guidance.References
1. Prasanna P, Patel J, Partovi S, et al. Radiomic features from the peritumoral brain parenchyma on treatment-naive multiparametric MR imaging predict long versus short-term survival in Glioblastoma Multiforme: Preliminary Findings. Eur Radiol. 2017;27(10):4188-4197.
2. Chaddad A, Kucharczyk M, Daniel P, et al. Radiomics in Glioblastoma: Current Status and Challenges Facing Clinical Implementation. Front. Oncol. 2019;9(374).
3. D’Alessio A, Proietti G, Sica G, et al. Pathological and Molecular Features of Glioblastoma and Its Peritumoral Tissue. Cancers. 2019;11(469).
4. Schmainda KM, Prah M. Data from Brain-Tumor-Progression. The Cancer Imaging Archive. 2019.
5. Jenkinson M, Smith SM. A global optimisation method for robust affine registration of brain images. Medical Image Analysis. 2001;5(2):143-156.
6. Yushkevich P, Piven J, Hazlett H, et al. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage. 2006;31(3)1116-28.
7. MATLAB, Deep Learning Toolbox and Feature Selection Toolbox 2018b. The MathWorks, Inc.
8. Griethuysen, JJM, Fedorov A, Parmar C, et al. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Research. 2017;77(21):104-107.
9. Emmert-Streib F, Dehmer M. High-Dimensional LASSO-Based Computational Regression Models: Regularization, Shrinkage, and Selection. Mach. Learn. Knowl. Extr. 2019;1(1):359-383.
10. Fradkin D, Muchnik I. Support Vector Machines for Classification. DIMACS Series in Discrete Mathematics and Theoretical Computer Science. 2000.
Figures