1728
Multiparametric MRI Texture Analysis in Prediction of Genetic Biomarkers in Patients with Brain Glioma1Radiology, Icahn School of Medicine at Mount Sinai, New York, NY, United States, 2Pathology, Icahn School of Medicine at Mount Sinai, New York, NY, United States, 3Radiology, Kaiser Permanente Fontana Medical Center, Fontana, CA, United States, 4Institute for Health Care Delivery Sciences, Icahn School of Medicine at Mount Sinai, New York, NY, United States, 5Neurology, Icahn School of Medicine at Mount Sinai, New York, NY, United States, 6Neurosurgery, Icahn School of Medicine at Mount Sinai, New York, NY, United States
Synopsis
In this retrospective study, we used a commercially available texture analysis software (Olea Medical) to construct a multiparametric MRI radiomic model that can be used to predict several important prognostic biomarkers in a cohort of patients with brain glioma. A total of 92 texture features were calculated from both FLAIR and T1C+ images using a volume-of-interest analysis encompassing the entire FLAIR hyperintense tumor. Radiomic features obtained from our multiparametric MR texture model were able to predict genetic biomarkers of brain glioma with predictive accuracies ranging from modest (62.4%) for MGMT to nearing 90% for IDH-1 and ARTX.
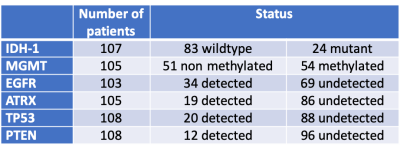
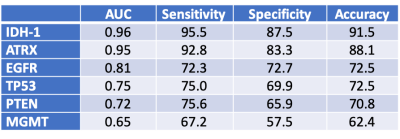
Methods: In this retrospective IRB approved study, patients were included if 1) had diagnosis of gliomas with known IDH-1, EGFR, MGMT, ATRX, TP53, and PTEN statuses from surgical pathology and 2) had preoperative MRI including FLAIR and T1c+. Tumor segmentation was performed by using volume-of-interest (VOI) analysis to include FLAIR hyperintense regions. VOIs were transferred for radiomic texture analysis obtained from FLAIR and T1c+ images using Olea Sphere software (Olea Medical). For each patient a total of 92 texture features including first-order, co-occurrence matrix, and run length matrix were calculated. Correlation between the biomarker statuses and texture features were assessed using Least Absolute Shrinkage and Selection Operator (LASSO) regression algorithm to prevent overfitting and increased interpretation. Receiver-operating characteristic (ROC) curve analysis was performed to determine the optimal parameters and threshold for predicting glioma prognostic biomarker statuses. Results: A total of 111 patients (age: 56 ± 15, M/F 66/45) were included. There were 21 patients with grade II-III gliomas and 90 patients with grade IV. The breakdown of genetic biomarkers is summarized in Figure 1. Figure 2 summarizes the AUC/sensitivity/specificity/accuracy measurements to predict each genetic marker from a combination of texture features that remained as significant contributors after logistic regression. The highest predictive performance was obtained for IDH-1 (AUC: 0.96, accuracy: 91.5%) from a combination of 6 features (informal correlation and maximum probability from FLAIR and cluster prominence, skewness, difference variance, and low gray level zone emphasis from T1c+). The second best performance was obtained for ATRX (AUC: 0.95, accuracy: 88.1%) from a combination of 11 features (gray level nonuniformity, cluster prominence, maximum probability, small area emphasis, dependence variance, and informal measure of correlation from FLAIR; and low gray level zone emphasis, matrix strength, high gray level emphasis, skewness, and difference variance from T1c+). The lowest predictive performance was obtained for MGMT methylation status (AUC: 0.65, accuracy: 62.4%) from a combination of 3 features (range and cluster prominence from FLAIR and interquartile range from T1c+).
Conclusion: Our multiparametric MR texture analysis can predict genetic biomarkers of brain glioma with good to excellent diagnostic accuracy nearing 90% for IDH-1 and ARTX, while only modest for MGMT.
Acknowledgements
No acknowledgement found.References
1. Kinoshita M, Sakai M, Arita H, et al. Introduction of high throughput magnetic resonance T2-weighted image texture analysis for WHO grade 2 and 3 gliomas. PLoS One. 2016 Oct 7;11(10):e0164268
2. Korfiatis P, Kline TL, Coufalova L, et al. MRI texture features as biomarkers to predict MGMT methylation status in glioblastomas. Med Phys. 2016 Jun;43(6):2835-2844. doi: 10.1118/1.4948668
3. Li Y, Liu X, Qian Z, et al. Genotype prediction of ATRX mutation in lower-grade gliomas using an MRI radiomics signature. Eur Radiol. 2018 Jul;28(7):2960-2968.