1726
The Diagnostic Performance of Multiparametric MRI Radiomics for Classification of Untreated Adult Gliomas1Department of Radiology Technology, Taibah University, Medina, Saudi Arabia, 2Department of Brain Repair & Rehabilitation, UCL, Queen Square, Institute of Neurology, London, United Kingdom, 3Department of Applied Health Research, UCL, London, United Kingdom, 4Lysholm Department of Neuroradiology, The National Hospital for Neurology and Neurosurgery, University College Hospitals NHS Trust, London, United Kingdom
Synopsis
This study aimed to assess the diagnostic performance of multiparametric MRI radiomics for glioma class prediction according to the WHO 2016 classification. Histogram features were extracted from prospectively acquired multiparametric MRI (pCASL, DSC-MRI, DCE-MRI, and DWI) in 32 patients with primary gliomas. The uncombined significant features of ASL, ADC, DSC, and DCE, revealed diagnostic performances varying from low (44% ) to fair (86%) and unable to predict all the histomolecular classes. However, combining them for each MRI method, independently, enhanced the diagnostic accuracy up to 100% and predict all the classes. This alludes the use of multimodal radiomics for glioma classification.
Introduction
Currently, histological biopsy is the gold standard for classifying gliomas according to the most recent histomolecular features. However, this process is both invasive and challenging when the lesion is at eloquent brain regions. Considering the interaction between the presence of the IDH-mutation, the upregulation of the hypoxia induced factor (HIF), angiogenesis and increased cellularity1, perfusion and diffusion MRI may be able to predict the presence of such mutations indirectly. Recently, several studies reported the subsidiary role of both perfusion and diffusion MRI2–5 in prediction of gliomas histomolecular class. However, the extracted values are often evaluated as a single value and mostly from the ‘hot-spot’ region of interest. In light of tumour heterogeneity, considering the signal intensity distribution in the segmented entire tumour volume via histogram analysis could enable more precise diagnosis free of inter-observer variability6 7. Useful, but “hidden” from the human eye information, can be extracted from the image using histogram texture analysis forming the basis of radiomics. This creates high dimensional data that can subsequently be reduced to ensure enhanced accuracy and elimination of redundant elements. This study aims to assess the diagnostic performance of multi-parametric MRI radiomics from the entire tumour volume using subsequently feature reduction and combination of the standalone modalities for classification of primary gliomas.Methods
Thirty-two adults with untreated gliomas were prospectively recruited and underwent multimodal MRI perfusion with pseudo-continuous arterial spin labelling (pCASL), dynamic susceptibility contrast-enhanced (DSC), and dynamic contrast-enhanced (DCE) MRI, as well as diffusion-weighted imaging (DWI). The gliomas were subsequently biopsied or surgically removed and the final diagnosis was established using the WHO 2016 classification. DCE maps were generated using both the modified Tofts model (mTK)8 and the Lawrence and Lee model (L&L)9. The entire tumour was segmented manually on FLAIR, while grey matter (GM) was used as an internal reference, being automatically segmented on a high-resolution T1-weighted volume. Histogram features (mean, standard deviation, 95th-percentile, kurtosis, skewness, median, inter-quartile range, mode, minimum, maximum, variance, entropy, median z-score, and slope of the cumulative distribution function) were extracted from absolute and relative (normalised) values (aT, rT, respectively). Additional percentiles were extracted from the ADC map, 10tile to 90tile with 10 increment. Based on the Kruskal-Wallis H test, only the statistically significant features were kept. These features were further reduced using pairwise correlation (PC) and backward elimination (BE). The final diagnostic performance was assessed using multinomial logistic regression, both for individual features as well as for combined features from each modality. The weighted average F1 (F) being used as indicator to reflect the classification accuracy for the imbalanced sample sizes.Results
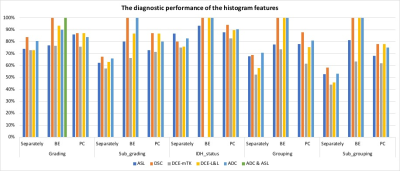
Figure 1 shows the diagnostic performance of the significant features separately and combined. The uncombined significant features of ASL, ADC, DSC, and DCE, revealed diagnostic performances varying from low (44% ) to fair (86%) and unable to predict all the histomolecular classes. However, linking the significant histogram features, mostly from rT, of each MRI method, independently, enhanced the diagnostic accuracy up to 100% and made it feasible to predict all the classes. As an individual technique, DSC showed superior diagnostic performance (overall 100%), though utilising both PCASL and ADC has similar accuracy (100%) to it. The DCE-L&L performed better than ASL (F range: 87% to 100%, 78% to 93%, respectively), but not the DCE-mTK (F range: 64% to 83). Notably, DCE-L&L attained identical diagnostic performance as DSC in term of the molecular classification.Discussion
Both ADC and DSC are widely used in current clinical glioma MRI protocols10. Based on our results, DSC might be omitted when using the combined histogram features from ADC while still achieving effective diagnoses. However, ADC does not provide information regarding the tumour’s hemodynamic nature. In contrast, though ASL has still not been fully adopted in routine glioma MRI scanning, our results revealed that the combined histogram features from PCASL and ADC had comparable diagnostic performance to DSC (overall 100%), confirming that DSC could be here safely skipped, something that would be important in light of the increasing awareness for gadolinium deposition, with the caveat of longer scanning time required for ASL and limited dissemination of PCASL. Nonetheless, out of the PCASL with multiple TIs acquired in 5min 9s in our study, only the ASL data at the 6th TI was considered for post-processing and analysis, rendering the actual PCASL scanning time about 1min 2s, similar to the DSC acquisition time (1min 26s). A critical step in radiomics for diagnostic purposes is feature reduction11. The reduction was performed here using PC and BE11,12, where the latter demonstrated better results. This might be because the PC method considers the relationship between both feature-class and feature-features, while the BE considers all the significant class-related features while ignoring feature-feature mutual information. These results, however, could be excessively optimistic, due to the small sample size, and larger cohorts are needed.Conclusion
Our preliminary investigation suggests that either DSC as an individual modality or alternatively combined ASL and ADC have similar and excellent accuracy as non-invasive biomarkers for gliomas classes prediction. This reinforces the evidence for the diagnostic accuracy of DSC and at the same time alludes to the use of multimodal radiomics for glioma classification.Acknowledgements
No acknowledgement found.References
1. Cairns RA, Harris I, Mccracken S, Mak TW. Cancer cell metabolism. Cold Spring Harb Symp Quant Biol. 2011;76(December 2011):299-311. doi:10.1101/sqb.2011.76.012856.
2. Brendle C, Hempel J-M, Schittenhelm J, et al. Glioma Grading and Determination of IDH Mutation Status and ATRX loss by DCE and ASL Perfusion. Clin Neuroradiol. 2017. doi:10.1007/s00062-017-0590-z. 3. Lin Y, Xing Z, She D, et al. IDH mutant and 1p/19q co-deleted oligodendrogliomas: tumor grade stratification using diffusion-, susceptibility-, and perfusion-weighted MRI. Neuroradiology. 2017;59(6):555-562. doi:10.1007/s00234-017-1839-6.
4. Xing XZ, Yang XX, She XD, Lin XY, Zhang XY, Cao XD. Noninvasive Assessment of IDH Mutational Status in World Health Organization Grade II and III Astrocytomas Using DWI and DSC-PWI Combined with Conventional MR Imaging. 2017.
5. Latysheva A, Emblem KE, Brandal P, et al. Dynamic susceptibility contrast and diffusion MR imaging identify oligodendroglioma as defined by the 2016 WHO classification for brain tumors: histogram analysis approach. Neuroradiology. 2019;61(5):545-555. doi:10.1007/s00234-019-02173-5.
6. Just N. Improving tumour heterogeneity MRI assessment with histograms. Br J Cancer. 2014;111(12):2205-2213. doi:10.1038/bjc.2014.512.
7. Zeng Q, Jiang B, Shi F, Ling C, Dong F, Zhang J. 3D pseudocontinuous arterial spin-labeling MR imaging in the preoperative evaluation of gliomas. Am J Neuroradiol. 2017;38(10):1876-1883. doi:10.3174/ajnr.A5299.
8. Tofts PS. I Modeling Tracer Kinetics in Dynamic. J Magn Reson Imaging. 1997;7(1):91-101.
9. Lawrence KS St., Lee T-Y. An Adiabatic Approximation to the Tissue Homogeneity Model for Water Exchange in the Brain: II. Experimental Validation. J Cereb Blood Flow Metab. 1998;18(12):1378-1385. doi:10.1097/00004647-199812000-00012.
10. Thust SC, Heiland S, Falini A, et al. Glioma imaging in Europe: A survey of 220 centres and recommendations for best clinical practice. Eur Radiol. 2018;28(8):3306-3317. doi:10.1007/s00330-018-5314-5.
11. Afshar P, Mohammadi A, Plataniotis KN, Oikonomou A, Benali H. From handcrafted to deep-learning-based cancer radiomics: Challenges and opportunities. IEEE Signal Process Mag. 2019;36(4):132-160. doi:10.1109/MSP.2019.2900993.
12. Hall MA, Smith LA. Correlation-based Filter Approach vs. Wrapper. 1995.