1723
Experience counts! Comparison of ROI placement strategies for radiomics analysis of gliomas1Department of Radiology, Leiden University Medical Center, Leiden, Netherlands, 2Department of Radiology, Kyoto Prefectural University of Medicine, Kyoto, Japan, 3Department of Radiology, University of Michigan, Ann Arbor, MI, United States
Synopsis
Glioma delineation is complex and time-consuming. We evaluated whether a technically less challenging method could potentially bypass this tedious process, when using a simple manual circular-ROI by less trained personnel. We assessed whether applying a circular-ROI to clinical 3T-MRIs from 12 glioma patients, would extract significantly different GLI features. We compared it to the gold-standard manual delineation approach, having 3 observers with different levels of expertise placing the ROIs. Experience matters to extract consistent GLI features across delineation runs. The different ROI methods extracted significantly different GLI features, emphasizing the importance of delineation strategies when analyzing heterogeneous tumors like gliomas.
Introduction
Gliomas comprise 81% of all brain malignancies(1). Radiomics, a novel field of image analysis, aims to retrieve quantitative imaging features to non-invasively assess tumors (2,3). For instance, gray level intensity (GLI) describes the distribution of voxel intensity within a region of interest (ROI) (4). Strict ROI delineation is believed to be necessary for reliable tumor quantitative analysis. However, manual delineation still being the current gold-standard approach, is highly time consuming, and given the diffuse nature of gliomas, extremely challenging (5). To date no study has shown whether the extraction of GLI distribution significantly differs upon a different and less arduous delineation strategy. To the best of our knowledge, an alternative and less laborious delineation strategy for gliomas has only been explored for radiotherapy purposes (6). To challenge the labor-intensive task of glioma delineation we assessed whether a more elementary ROI strategy could detect the same GLI distribution. Additionally, we also determined whether experience matters for this task. If experience does not substantially matter, burdensome and expensive work can be relieved from overloaded clinicians.Methods
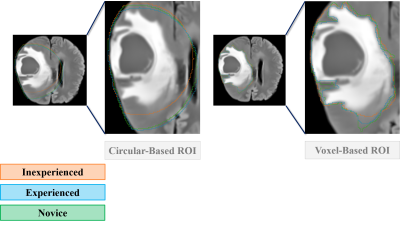
This is a retrospective study including clinical MR images (FLAIR, T2W and ADC maps) from a total of 12 glioma patients (WHO grades II = 5; III = 3; IV = 4) scanned at 3 Tesla (Skyra, Siemens Healthineers, Erlangen, Germany). All images were rigidly co-registered to the contrast enhanced T1W image. Given the exploratory character of our study, 3 representative slices from each tumor were selected on the FLAIR image. Our strict ROI obeyed the current clinical gold practice in which a voxel-based delineation is considered. The broad ROI was defined by establishing a center of gravity on the axial plane on the FLAIR image of each tumor, for clear edema visualization, and expanding a circular ROI covering the least healthy tissue (Figure 1). Each ROI delineation was performed twice by each observer and posteriorly transferred to the remaining MR images for feature extraction. Features were retrieved from Pyradiomics® open-source python package, fully described in: (4). 3D-Slicer software graphical user interface (GUI) was used for tumor delineation and feature extraction. We performed a two-sided Wilcoxon signed-rank test to assess whether the magnitude of the difference between the quantitative features extracted within each delineation strategy differed significantly. A non-parametric test was applied given the small sample size. The features were considered to significantly differ when the p-value was below 0.05. Correction for multiple comparisons applied. In addition, to assess the observers’ experience and account for the manual nature of the delineation strategies, we calculated the intra-class correlation coefficient (ICC) between the features retrieved from both ROI strategies among the 3 observers and within each observer, respectively. Confidence interval was set at 95% with significance level at .01. Statistical analysis was done in SPSS (IBM SPSS - Windows, V25.0).Results
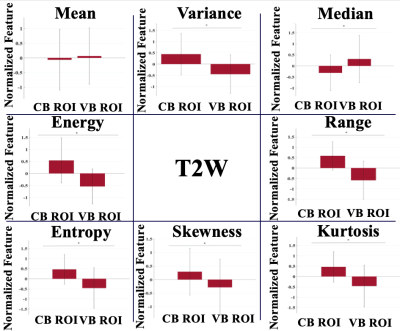
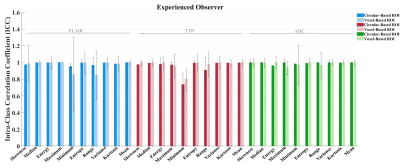
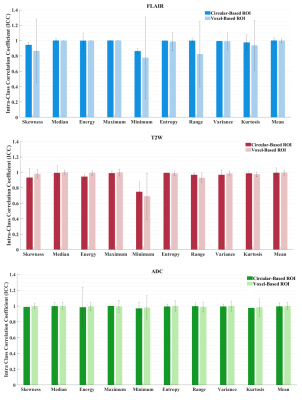
We found that overall reproducibility of the GLI features, between three independent observers was good (ICC > 0.7; 1 = total agreement) for the circular-based and voxel-based ROIs in both delineations runs (Figure 4). These results suggest that overall clinical experience does not have a significant influence when the ROI is placed for voxel intensity assessment. However, we found a higher intra-observer feature agreement from the delineations performed by the experienced observer for both broad and strict delineation strategies (Figure 3). Moreover, our results indicate that overall, the delineation method applied for ROI placement extract significantly different (p < 0.05) GLI features (results not shown). Exceptions were found for the mean GLI extracted from the T2W image (Figure 2).Discussion
In this study we confirmed that the level of the observer’s experience for ROI delineation in gliomas does not significantly impact the extraction of MR image voxel GLI distribution, given a high inter-observer agreement for each quantitative feature (7). On the other hand, observers experience seems to matter for a steady ROI placement regarding the extraction of the same features. Experience, most likely through acquired training, allows for a more consistent assessment of tumor boundaries, and consequent ROI delineation. This suggests that this burdensome task can be taken over by less experienced personel given the right training and learning procedures. Our circular based whole tumor delineation strategy yielded significantly different GLI features from voxel-based ROI. This can be possibly explained by the surrounding healthy tissue included (8). Given the fundamental differences between healthy and tumorous tissue, image intensity is expected to appear within different intensity ranges (9). According to our results, the delineation strategy has a significant impact for GLI extraction of clinical MR images.Conclusion
Circular based delineation method extracts different tissue based on significantly different GLI values detected. Thorough delineations of gliomas seems to be necessary for GLI assessment of clinical MR images. Reliable quantitative features can be extracted from whole tumor ROIs delineated by less experienced observers relieving time consuming work from overloaded clinicians, upon proper training. We encourage future studies to further explore alternative ROI delineation methods in additional radiomics features. Larger sample sizes and distinction between high-grade and low-grade gliomas should also be considered.Acknowledgements
No acknowledgement found.References
1. Ostrom QT, Bauchet L, Davis FG, Deltour I, Fisher JL, Langer CE, et al. The epidemiology of glioma in adults: a "state of the science" review. Neuro Oncol. 2014;16(7):896-913.
2. Kickingereder P, Neuberger U, Bonekamp D, Piechotta PL, Gotz M, Wick A, et al. Radiomic subtyping improves disease stratification beyond key molecular, clinical, and standard imaging characteristics in patients with glioblastoma. Neuro Oncol. 2018;20(6):848-57.
3. Thust SC, Heiland S, Falini A, Jager HR, Waldman AD, Sundgren PC, et al. Glioma imaging in Europe: A survey of 220 centres and recommendations for best clinical practice. Eur Radiol. 2018;28(8):3306-17.
4. van Griethuysen, J. J. M., Fedorov, A., Parmar, C., Hosny, A., Aucoin, N., Narayan, V., Beets-Tan, R. G. H., Fillon-Robin, J. C., Pieper, S., Aerts, H. J. W. L. (2017). Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Research, 77(21), e104–e107.
5. Weller M WW, Aldape K, Brada M, Berger M, Pfister S;, Nishikawa R, Rosenthal M, Wen PY, Stupp R, Reifenberger G. Glioma. Nature Reviews. 2015;1.
6. Guo L, Wang P, Sun R, Yang C, Zhang N, Guo Y, et al. A fuzzy feature fusion method for autosegmentation of gliomas with multi-modality diffusion and perfusion magnetic resonance images in radiotherapy. Sci Rep. 2018;8(1):3231.
7. Abdallah MB BM, Wantz-Mézières, Gaudeau Y, Taillandier L, Moureaux JM. Statistical evaluation of manual segmentation of a diffuse low-grade glioma MRI dataset. IEEE. 2016:4403-6.
8. Papanikolaou N SJ. An Introduction to Radiomics: Capturing Tumour Biology in Space and Time. Oncologic Imaging REVIEW. 2018;3(1):61-71
9. Kao HW, Chiang SW, Chung HW, Tsai FY, Chen CY. Advanced MR imaging of gliomas: an update. Biomed Res Int. 2013;2013:970586
Figures