1722
MRI-based radiomics analysis for differentiating subtypes of glioma1Lanzhou University Second Hospital, Lanzhou, China, 2Huiying Medical Technology Co., Ltd., Beijing, China
Synopsis
Radiomics provides a tool for comprehensive quantification and visualization of intra-tumor heterogeneity at the radiological level. Several radiomics studies have been reported in prediction of the survival and treatment response of glioma. And most researches had focused on binary classification of the Low-grade glioma (grade II) and high-grade glioma (grade III and grade IV). However, the influence of different MRI scan plane on the radiomic features of glioma has not been investigated. The purpose of this retrospective study was to demonstrate the feasibility of radiomics methods to determine the three subtypes (grade II, III, and IV) of glioma based on multi-sequences and different scan plane of magnetic resonance imaging (MRI).
Introduction
Gliomas are the most aggressive primary brain tumors arising from glial cells and present poor survival rates[1-2]. They are divided into four categories (grades I, II, III and IV) by the World Health Organization grading system on the basis of the invasively histopathology. Although molecular markers may play an important role in determining the individualizing treatment and accurately estimating prognosis, the classification of glioma patients can’t proceed on the basis of molecular characteristics alone [3-4]. Radiomics has emerged as a powerful methodology to quantify the characteristics of tumors in recent years, bringing a novel biomarkers and new hope for the glioma grading in a non-invasive manner [5-6]. In this study, we aimed to construct the radiomics model to diagnose the three subtypes (grade II, grade III, and grade IV) of glioma non-invasively based on the conventional multi-MR sequences (T2WI and 3D-T1WI-CE), and investigated the influence of the different MRI scan plane.Materials and Methods
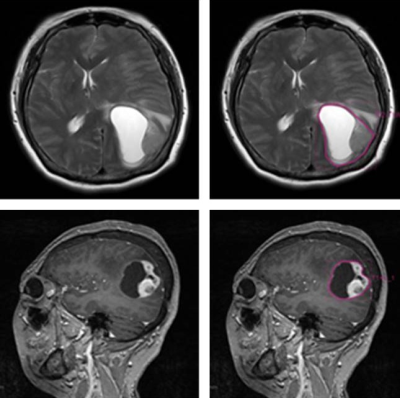
This retrospective study enrolled 208 patients (with grade II 64, grade III 60, and grade IV 84) who were pathologically confirmed as primary glioma in LanZhou University Second Hospital between Mar 2016 and Nov 2018. Patients’ age and sex were treated as control variables. The MR images of 208 patients were collected from the Siemens Verio 3T MR scanner with a 8-channel head coil. The slice thickness of all T2WI sequences was 5.5mm and the field of vision (FOV) was 257×230 mm. The slice thickness of all sagittal isotropical T1WI-CE sequences was 1 mm and the field of vision (FOV) was 286×256 mm. To obtain volume of interest (VOI) for further analysis, all the data were uploaded to the Radcloud platform (http://mics.radcloud.cn/). The VOI of glioma was the tumor core region manually defined by two experienced radiologists with overall consideration of image features of two sequences, and manually delineated by the junior one. Figure 1 showed an example of the manual segmentation. After VOI segmentation, 1029 radiomic features were extracted from VOI including tumor image intensity, shape and size features, and texture features in each MRI sequences. Next, the features were standardized to eliminate the adverse effects caused by the singular sample data. In order to reduce the redundancy of features and improve the robustness of results, the variance threshold, K best and least absolute shrinkage and selection operator (LASSO) algorithm was adopted to select the features in each single MRI sequence and the combined sequence T1WI+T2WI, respectively. Then three radiomics-based models were constructed with the selected features of each single sequences and the joint sequences respectively on the training dataset for the glioma subtypes classification. The five-fold cross-validation method and support vector machine (SVM) algorithms were utilized for radiomics-based model constructing. And the diagnostic performance of the models were verified on the validation dataset, and were evaluated by receiver operating characteristic curves with indicators of sensitivity, specificity.Results
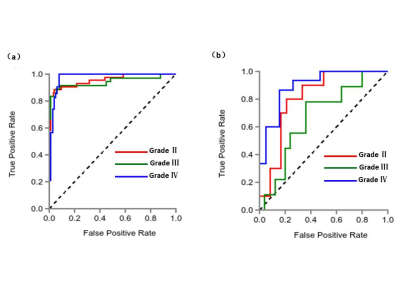
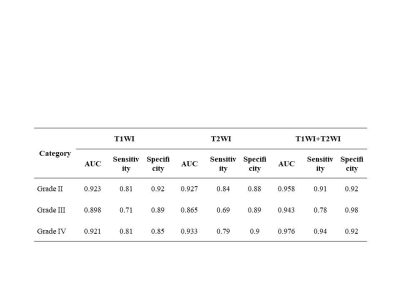
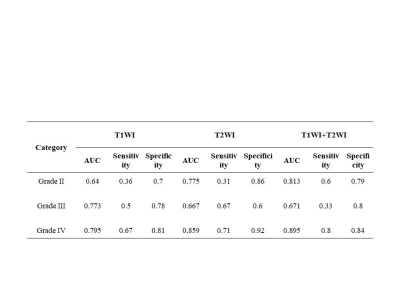
After feature selection by LASSO method, the numbers of remaining features of T2WI, T1WI-CE and the combined T1WI+T2WI were 27, 14 and 31, respectively. The results including ROC related AUC value, sensitivity, and specificity of the three subtypes grade II, grade III, and grade IV, of the three radiomics-based models under SVM classifier respectively were summarized in Table 1 and Table 2. Table 1 showed the results on training dataset, and Table 2 showed the results on validation dataset. As we can see, the diagnosis performance on grade II and grade IV was better than that on grade III, and the combination of two sequences had the better diagnosis performance with AUC (grade II 0.958, grade III 0.943 and grade IV 0.977 in training dataset, and grade II 0.813, grade III 0.617 and grade IV 0.895 in validation dataset). Figure 2 showed the ROC curve of the combined sequence T1WI+T2WI on training dataset and validation dataset.Discussion and Conclusion
In this study we evaluated the glioma’s subtypes using multi-sequences and different scan plane of MRI based on radiomics, and the subtype classification of grade III and grade IV has been investigated for the first time. The results showed that the radiomics-based models had high potential for diagnosis the subtypes of glioma preoperatively. These models were expected to assist clinicians to make better clinical diagnosis and treatment strategies, and which indicated that radiomics enabled to accelerate the development of personalized medicine.Acknowledgements
1.The National Natural Science Foundation of China ( approve number: 81960309).
2.The Natural science foundation of gansu province,China(approve number:18JR3RA317).
3.Huiying Medical Technology Co., Ltd., Beijing, China.
References
[1] Vamvakas A, Williams SC, Theodorou K, et al. Imaging biomarker analysis of advanced multiparametric MRI for glioma grading[J].Phys Med. 2019,60:188-198.
[2] Hao Z, Ken C, Harrison X. B,et al. Machine learning reveals multimodal MRI patterns predictive of isocitrate dehydrogenase and 1p/19q status in diffuse low and high-grade gliomas [J].J Neurooncol. 2019,142(2): 299–307.
[3] Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary [J]. Acta Neuropathol. 2016, 131(6): 803-820.
[4] Yip SS, Aerts HJ. Applications and limitations of radiomics [J].Phys Med Biol. 2016, 61(13): R150-R166.
[5] Yan T, Wei M, Xiao-chun W, et al.Improving survival prediction of high-grade glioma via machine learning techniques based on MRI radiomic, genetic and clinical risk factors [J].Eur J Radiol. 2019,120:108609.
[6] Chaddad A, Kucharczyk MJ, Daniel P, et al. Radiomics in Glioblastoma: Current Status and Challenges Facing Clinical Implementation.[J].Front Oncol. 2019,9:374.
Figures