1707
Analysis of Accuracy and Precision of Recommended Protocols for Dynamic Susceptibility Contrast MRI for Brain Metastases
Natenael B Semmineh1, Usha Bhalla2, Laura C Bell1, Ashley M Stokes1, Matthew D Lee3, Leland Hu4, Jerrold L Boxerman3,5, and C Chad Quarles1
1Division of Neuroimaging Research, Barrow Neurological Institute, Phoenix, AZ, United States, 2Brown University, Providence, RI, United States, 3Alpert Medical School - Brown University, Providence, RI, United States, 4Department of Radiology, Mayo Clinic Arizona, phoenix, AZ, United States, 5Department of Diagnostic Imaging, Rhode Island Hospital, Providence, RI, United States
1Division of Neuroimaging Research, Barrow Neurological Institute, Phoenix, AZ, United States, 2Brown University, Providence, RI, United States, 3Alpert Medical School - Brown University, Providence, RI, United States, 4Department of Radiology, Mayo Clinic Arizona, phoenix, AZ, United States, 5Department of Diagnostic Imaging, Rhode Island Hospital, Providence, RI, United States
Synopsis
Numerous methods have been proposed to overcome the confounding influence of contrast agent leakage on DSC-MRI derived CBV maps in brain tumors. Recently, we leveraged a digital reference object to identify the optimal acquisition and analysis methods for GBM patients, a protocol that is now being recommended as the standardized approach for all clinical scans. It is unknown if the biological differences between GBM and metastases would confound the use of this protocol in patients with brain metastases. In this study we characterize the influence of this GBM-centric protocol resulted in lower metastases CBV accuracy and precision.
Introduction
DSC-MRI derived cerebral blood volume (CBV) maps are widely used for brain tumor patient management [1]. However, the accuracy of CBV maps depends heavily on the acquisition parameters. We recently developed and leveraged an in silico digital reference object (DRO) to aid in identifying optimal protocols for glioma patients [2]. This protocol is now being recommended as the standardized DSC-MRI approach for all clinical scans. It is currently unknown if the biological differences between glial and non-glial tumors could confound the fidelity of this recommended protocol. The goal of this study is to determine the accuracy and precision of the optimized glioma DSC-MRI protocols when applied to the biological conditions found in brain metastases.Methods
The DRO was founded using a validated computational strategy and includes MRI signals for realistic 3D tissue structures modeled using ellipsoids (cells) packed around randomly oriented cylinders (vessels) and accounts forthe static magnetic field strength, inter-compartment susceptibility differences, the water proton diffusion coefficient, and pulse sequence parameters [2,3]. To ensure that the DRO’s simulated DSC-MRI signals accurately represent the magnitude and distribution of CA-induced T1and T2* changes observed across typical metastases brain tumor, we used voxel-wise patient data to train the DRO to identify the relevant structural features. The training dataset included patient data from 20 metastases (17,077 voxels), which was acquired using TE=40ms, TR=1.29s, flip-angle=60o, with a dosing scheme of quarter-dose preload followed by full-dose injection of Gd-BT-DO3Aat 1.5T. To ensure clinical relevance the DRO’s input tissue structure (e.g. cell size, shape) and physical parameters (e.g. pre-contrast T1) were systematically permutated until the distribution of percent signal recovery (PSR) values, a parameter that reflects CA-induced T1 and T2* changes, and their mean and standard deviation across all the simulated signals closely matched those in the training dataset. Further,the DRO was validated by comparison with a dataset acquired in a separate small cohort of metastases (n = 6) patients for pulse sequences and contrast agent dosing schemes different from those in the training dataset. To investigate the accuracy and precision of the optimized glioma DSC-MRI protocols on the reliability of CBV measurements in brain metastases, we compared tumor CBV simulated with and without (Ktrans = 0) CA leakage effects for two optimized glioma DSC-MRI protocols, Method 1: acquired with full dose followed by full dose at intermediate flip-angle (60o), 30 ms TE and 1.5 s TR and Method 2: acquired with no preload followed by full dose dosing scheme at low flip-angle (30o), 30 ms TE and 1.5 s TR.Results and Discussion
Figure 1A shows the agreement between the in vivoand simulated mean and standard deviation of PSR for metastases tumor in the training dataset. Figure 1Bcompares simulated and in vivoPSR distribution for the validation datasets. The DRO captured the full range of PSR values for metastases data acquired using TE=24ms, TR=1.8s and flip angle=90o. The results indicate that the trained DRO can accurately model the full range of CA-induced T1 and T2* effects for different sets of pulse sequence parameters and CA dosing schemes. Figure 2 shows the scatter and Bland-Altman plots for metastases and GBMs at both protocols. As shown in Figure 2A-B, Method 1 resulted in a CCC and CV of 0.98 and 6.8% for GBMs, and 0.84 and 14.5% for metastases, respectively. The second protocol yielded a CCC and CV of 0.92 and 7.5% for GBMs, and 0.86 and 13.6% for metastases, Figure 2C-D. The data shows that the protocols tend to result in underestimated CBV values, as shown in the Bland-Altman plots. These preliminary results indicates that both protocols optimized for GBMs yielded less accurate and less precise CBV maps for metastases, due to the difference in the magnitude of T1and T2* contrast agent leakage effects and the effective interplay of the leakage correction method and imaging protocol to reduce this effects.Conclusion
Although the accuracy and precision of the glioma-optimized protocols are not as robust in brain metastases, it is important to note that a CCC of 0.86 or 0.84 is reasonable and unlikely to undermine the clinical utility. Of particular note, the higher CV values could influence the interpretation of CBV values across time and influence its use in clinical trials, encouraging the use of an optimized protocol in trials focused on metastases. We are currently expanding this investigation for primary central nervous system lymphoma, given that it also exhibits highly dissimilar biological characteristics. Future work will seek to determine optimal protocols for metastases and lymphomas, which could aid in the application of DSC-MRI in patient and clinical trial use.Acknowledgements
NCI R01 CA213158-01References
- Boxerman JL, Schmainda KM, Weisskoff RM. Relative cerebral blood volume maps corrected for contrast agent extravasation significantly correlate with glioma tumor grade, whereas uncorrected maps do not. AJNR Am J Neuroradiol. 2006;27:859–867.
- Semmineh N, Bell L, Stokes A, Hu L, Boxerman J, Quarles C. Optimization of acquisition and analysis methods for clinical dynamic susceptibility contrast (DSC) MRI using a population-based digital reference object. AJNR Am J Neuroradiol. 2018;39:1981–1988.
- Semmineh NB, Xu J, Boxerman JL, Delaney GW, Cleary PW, Gore JC, et al. An efficient computational approach to characterize DSC-MRI signals arising from three-dimensional heterogeneous tissue structures. PLoS One 2014;9. doi:10.1371/journal.pone.0084764.
Figures
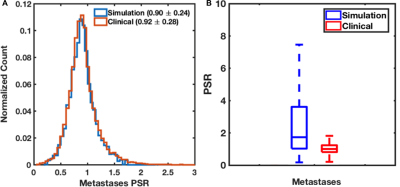
Figure 1: (A) A database of DSC-MRI of 20 metastases patients (17,077 voxels) were used to train input parameter permutations for the DRO, ensuring that the PSR distributions in vivoand in silicoare in strong agreement. (B) Validation of the DRO. The PSR distributions in the in vivovalidation dataset are a subset of those found in the DRO using two sets of pulse sequence parameters. Note that the DRO is not expected to precisely match the PSR distribution in the validation data, only the range of values, because it was trained using a much larger, more heterogeneous database.
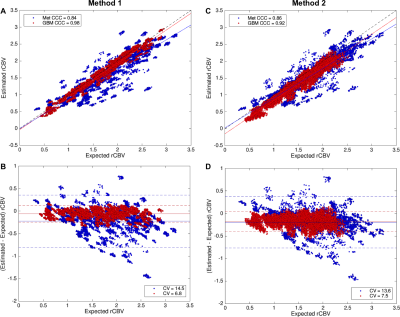
Figure 2. Scatter and Bland-Altman plots comparing CBV estimates for [1+1] dosing scheme using intermediate flip angle (A-B), and or [0+1] dosing scheme using low flip angle (C-D), For GBM (red) metastases(blue). Both protocols yielded lower accuracy and precision for metastasesas compared to GBM.