1694
Differentiating lymphoma from grade-IV glioma using susceptibility weighted imaging based texture feature at 3T MRI1Center for Biomedical Engineering, Indian Institute of Technology Delhi, New Delhi, India, 2Philips Health Systems, Philips India Limited, Gurugram, India, 3Department of Radiology, Fortis Memorial Research Institute, Gurugram, India, 4Department of Neurosurgery, Fortis Memorial research Institute, Gurugram, India, 5Department of Biomedical Engineering, All India Institute of Medical Sciences, New Delhi, India
Synopsis
No standalone method is yet reported to differentiate PCNSL and grade-IV glioma with highest accuracy and diagnostic confidence. Imaging based differentiation between these two types has been a challenging problem. Objective of this study is to evaluate performance of texture-based features of SWI in improved differentiation of PCNSL from grade-IV gliomas. Proposed approach based upon texture feature-extraction from segmented SWI lesion, enabled automatic classification of tumors into primary central nervous system lymphoma and grade-IV glioma cases. One of the texture feature, Contrast, provided highest AUC along with high sensitivity and specificity. This classification might improve diagnosis and grading of tumors.
Introduction:
Primary central nervous system lymphoma (PCNSL) is a common type of malignant tumor, which is different from glioma in terms of tumor biology as well as treatment planning. Mostly PCNSL treatment involves chemo and/or radio-therapy [1-2], whereas surgical resection is the most common practice in case of grade IV gliomas for survival improvement. Imaging based differentiation between these two types has been a challenging problem. ADC, T1-post contrast, FLAIR, DTI, DCE-perfusion MR sequences have been shown to differentiate up to certain accuracy [3]. No standalone method is yet reported to differentiate PCNSL and grade-IV glioma with highest accuracy and diagnostic confidence. Ding et.al have reported use of susceptibility weighted imaging (SWI) in classifying lymphoma based on the lower incidence of hemorrhages and vessels compared to glioma and metastases, with an accuracy better than conventional T1-post contrast or DWI imaging methods alone [4]. Texture based feature extraction methods are also being employed as one of the ways for differentiation. Leon et.al and few other studies have already reported that texture features of T1-post contrast sequence differs in lymphoma compared to GBM [5]. Objective of this study is to evaluate performance of texture-based features of SWI in improved differentiation of PCNSL from grade-IV gliomas.Material and Methods
This retrospective study, approved by IRB, included total 38 patients (15 PCNSL and 23 grade IV glioma confirmed histologically as per WHO-2016-criteria). These patients underwent brain MRI on a 3.0T scanner (Ingenia, Philips Healthcare, The Netherlands). SWI data were acquired with 4 echoes at 5.6, 11.8, 18, 24.2ms (TR=31ms; flip-angle=17°, slice-thickness=1.0mm, acquisition-matrix=384×384, FOV=240×240mm2). The SW-Magnitude images were obtained from scanner by multiplying fast-field-echo (FFE)-M image with a phase-mask derived from PADRE (Phase-Difference-Enhanced-imaging) filtering process. The structural T2, FLAIR and DWI weighted images were also obtained and used to define the tumor boundary manually and were co-registered with the SWI magnitude images. For each tumor, polygonal ROIs were manually marked on registered FLAIR hyper-intense regions visible in all the tumor-containing slices and the ROIs were copied to subsequent SWI images. ROI were marked by PhD student and verified by experienced radiologist. Lesion area was segmented by MATLAB-based in-house developed algorithm. The segmented lesion intensities were normalized with average intensity in the contralateral normal area. In this study we computed second-order texture features from multidimensional histograms: the mean of the 13 Haralick features computed from the gray-scale co-occurrence matrix (GLCM). The GLCM functions characterize the texture of an image by calculating how often pairs of pixel with specific values and in a specified spatial relationship occur in an image, creating a GLCM, and then extracting statistical measures from this matrix. GLCM considers the spatial relationship of the pixels taken over all 13 neighbor orientations: Correlation, Energy, Contrast, Sum of squares variance, Entropy, Homogeneity, Sum average, Sum entropy, Sum variance, Difference entropy, Difference variance, Information measure of correlation1, and Information measure of correlation2. Statistical unpaired-t-test was performed to check whether there is any significant difference of GLCM based metrics between PCNSL and grade-IV glioma. ROC curve analysis was used to evaluate the performance of GLCM based texture feature between pCNSL and grade-IV glioma by comparing the AUC. The highest sum of sensitivity and specificity was used to establish the cutoff values in differentiating pCNSL and grade-IV glioma. All statistical analysis were performed using MedCalc [6]. p ≤ 0.05 indicated a statistically significant difference.Results and Discussion
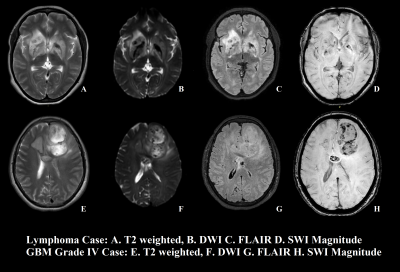
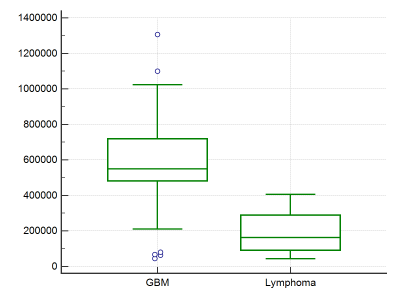
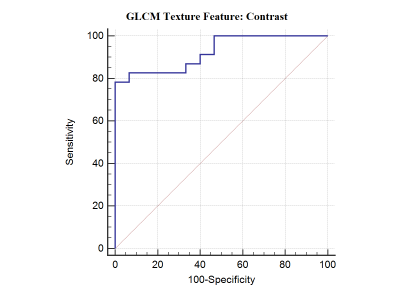
Figure 1 shows sample case of each lymphoma and Glioma grade-IV.Unpaired-t-test of 13 features provided significant p value (p<0.05) only for GLCM texture feature “Contrast”. Figure-1 summarizes the GLCM: contrast parameter for PCNSL and Glioma grade-IV. Figure 2 demonstrates the results of the ROC curve analysis where the AUC is proportional to the diagnostic accuracy. GLCM: contrast parameter showed an AUC of 0.925, for the discrimination of pCNSL and grade-IV glioma. The best cut-off value of Contrast texture feature obtained from the ROC analysis is >393862. This cut-off can differentiate between pCNSL and grade-IV glioma with a 82.61% and specificity of 92.33%. This study demonstrate GLCM-Contrast values were much higher in Grade-IV compared to primary lymphoma. This feature defers the calculation of the intensity contrast-linking pixel and its neighbor over the whole Lesion area/region. Contrast is a measure of the local variations present in an image.
In grade-IV glioma, lesion-segmented region in SWI has a large amount of variation compared to lymphoma, due to higher amount of hemorrhage and vessels presence inherently. Thus Contrast texture feature captured this information and provided differentiation between these two types of lesions. This approach, is more objective and quantitatively differentiates lymphoma from grade-IV glioma. Further studies on more number of patients and use of additional features for improving accuracy of differentiation. In the current study, SWI image intensities were normalized with values in the contralateral tissue. This normalization takes care of any intensity-scaling bias that could be present in the datasets.
Conclusion
Proposed approach based upon texture feature extraction from segmented SWI lesion, enabled automatic classification of tumors into primary central nervous system lymphoma and grade-IV glioma cases. One of the texture feature, Contrast, provided highest AUC along with high sensitivity and specificity. This classification might improve diagnosis and grading of tumors.Acknowledgements
No acknowledgement found.References
[1] Shiels MS, Pfeiffer RM, Besson C, et al.: Trends in primary central nervous system lymphoma incidence and survival in the U.S. Br J Haematol 2016; 174: 417–24.
[2] Kuker W, Nagele T, Korfel A, et al.: Primary central nervous system lymphomas (PCNSL): MRI features at presentation in 100 patients. J Neurooncol 2005; 72: 169–77.
[3] Zacharia TT, Law M, Naidich TP, Leeds NE:Central nervous systemlymphoma characterization by diffusion-weighted imaging and MRspectroscopy.J Neuroimaging2008,18:411–17.
[4] Y. Ding, Z. Xing, B. Liu, et al., Differentiation of primary central nervous system lymphoma from high-grade glioma and brain metastases using susceptibility weighted imaging, Brain Behav. 4 (2014) 841–849.
[5] Alcaide-Leon P, Dufort P, Geraldo AF, et al. Differentiation of Enhancing Glioma and Primary Central Nervous System Lymphoma by Texture-Based Machine Learning. AJNR Am J Neuroradiol 2017;38:1145-50. [6] MedCalc Statistical Software Version 14.8.1 (2014) MedCalc Software, Ostend, Belgium.