1577
Ex-vivo Imaging of Intracerebral Fetal Neural Transplants in a Huntington Patient at 7T: Graft Survival and Interconnectivity 11 year post-OP
David Lohr1, Philipp Capetian2, Bernhard Landwehrmeyer3, and Laura Maria Schreiber1
1Chair of Cellular and Molecular Imaging, Comprehensive Heart Failure Center (CHFC), University Hospital Würzburg, Würzburg, Germany, 2Department of Neurology, University Hospital Würzburg, Würzburg, Germany, 3Department of Neurology, University Hospital Ulm, Ulm, Germany
1Chair of Cellular and Molecular Imaging, Comprehensive Heart Failure Center (CHFC), University Hospital Würzburg, Würzburg, Germany, 2Department of Neurology, University Hospital Würzburg, Würzburg, Germany, 3Department of Neurology, University Hospital Ulm, Ulm, Germany
Synopsis
The brain of a Huntington Disease (HD) patient who received neural fetal tissue transplants 11 years before her death, was analyzed ex-vivo in a 7T MRI scanner with a total scan time of 25 hours and 8 minutes. Signs of engrafted tissue could be found on all sites which received a stereotactic implantation (caudate nucleus, putamen) with a tendency to overgrowth for the right hemisphere. Deterministic fiber tracking based on DTI images clearly demonstrated interconnectivity of individual grafts to numerous brain regions.
Introduction
Replacing neurons in circumscribed brain regions lost through neurodegenerative disease (such as HD) is the goal of neuro-transplantation. Since adult neurons are post mitotic and too sensitive to survive grafting procedures, cns tissue of aborted fetus has usually been employed. Clear proof of clinical benefit achieved by this procedure is still lacking. Engraftment into the host brain circuit and long-term survival of this allogenic material is a prerequisite of clinical success. Connection to other brain regions over long distances is hard to prove in histological specimens of human brains, but a particular strength of diffusion tensor imaging (DTI). Therefore, we chose to analyze a brain from a HD patient, who received fetal neural transplants 11 years before her death, using ex-vivo ultra-high resolution MRI at 7T.Methods
The HD patient was included in the MIG-HD study (approved by the local ethics committee) after giving informed consent: Tissue of the whole ganglionic eminence from mechanically aborted fetus was collected, minced into small pieces and stereotactically implanted into the caudate nucleus and putamen by two operations, one hemisphere each. After the death of the patient by pneumonia in end-stage HD, the brain was removed during autopsy, fixed, and kept in Formalin for several months until further analysis.The brain was placed in a plastic container and the sample position fixed using Fomblin (YL VAC 06/06, Solvay Specialty Polymers Italy SpA) soaked sponges (Figure 1a). The container was then slowly filled with Fomblin and excess air removed from the sponges and the brain using a vacuum desiccator (Figure 1b).
Measurements were performed on a 7T whole-body MRI system (Siemens MAGNETOM™ Terra) using a 1Tx/32Rx head coil (Figure 1c).
Using a 3D sequence we acquired SWI data with ultrahigh spatial resolution for the whole brain (isotropic resolution: 0.25mm). Measurement parameters were TE: 8.85ms, TR: 38ms, slices: 448,FOV: 200mm, FOVphase: 66%, R:3 (GRAPPA), bw:200 Hz/Px averages: 32, total scan time: 16h20m.
Another SWI scan was used for ultra-high in-plane resolution (0.15mm). Parameters were TE: 5.87ms, bw: 400Hz/Px, slice thickness: 0.5mm, averages: 12, rest: see isotropic SWI acquisition.
DTI data was collected with isotropic 1.0mm resolution for the whole brain. A Stejskal-Tanner diffusion preparation was used for 2 shells (10 averages, 30 directions MDDW b1500 and 64 directions MDDW b2500). Reference images (n=64) were collected for both shells and with reversed phase encoding for proper post-processing. Other parameters were TE: 70ms, TR: 13500ms, 120 slices, FOV:200mm, FOVphase: 78%, R=3 (GRAPPA), total scan time: 4h20min.
Results
Figure 2 shows representative SWI images and corresponding SNR for both acquisitions (isotropic and high in-plane resolution). Figure 3 shows a coronal section at the level of the anterior striatum with surviving graft tissue (red) at the end of the stereotactic trajectories (green). The iron rich, degenerated striatum gave an excellent contrast to the predominantly intact grafts on both sides. The size of the grafts differed more between hemispheres than between transplantation sites. Defining such a graft as seeding point in the DTI sequences demonstrated widespread fibers connections to numerous regions of the brain, but mainly the brain stem (Figure. 4).Discussion
After almost three decades of performing neurotransplantation in HD patients, results from large, systematic trials are still lacking with the biggest one (MIG-HD) still ongoing. New cell sources appearing on the horizon (e.g. neurons from induced pluripotent stem cells) might become available in the near future, but the question remains, if neural stem cells and neuronal precursors are able to successfully engraft in the host brain, integrate in neuronal circuits and survive for many years.However, integration into the host circuits and the connection to distant brain sites is almost impossible to prove histologically in post-mortem human brain samples. Therefore, we chose to employ the superior imaging possibilities of a 7T MRI scanner with the long scanning times possible only fort post-mortem material. To our knowledge, this represents the first attempt of post-mortem MRI imaging of neural transplants in a human brain.
A workstation with Intel(R) Core™ i7-4790 CPU and 32GB RAM proved insufficient for processing of the DTI data volume. We are currently implementing a post-processing pipeline using dedicated hardware.
We found the best contrast between the iron rich degenerating striatum in HD and the obviously still intact grafts was achieved using SWI sequences. They allowed us to define seeding points within the grafts that demonstrated widespread fiber connections. Naturally, it is not possible to distinguish afferent and efferent connections and rule out fibers merely passing through the grafts. However, the assumption that neural grafts in the human brain are not integrating in the host circuit seems unfounded to us.
Further measurements are required in other patients to demonstrate the general findings we made in this pilot study.
Conclusion
Our post-mortem imaging protocol demonstrated long-term survival of grafted fetal tissue in a HD patient brain despite widespread degeneration of the host striatum. DTI sequences demonstrated widespread interconnection of individual grafts with distant brain regions.Acknowledgements
Financial support: German Ministry of Education and Research (BMBF, grant: 01E1O1504).References
No reference found.Figures
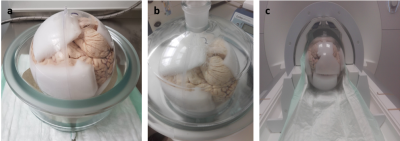
Figure 1: Sample preparation. a: Setup in vacuum desiccator. Ascending bubbles indicate successful removal of air. b: Placement in plastic sphere with sample positioning fixed using Fomblin soaked sponges. c: Placement in the head coil, measurement position.
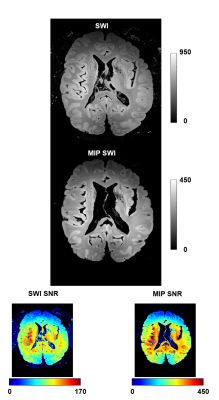
Figure 2: SWI and MIP images with respective SNR maps for the high resolution (in-plane) SWI scan.
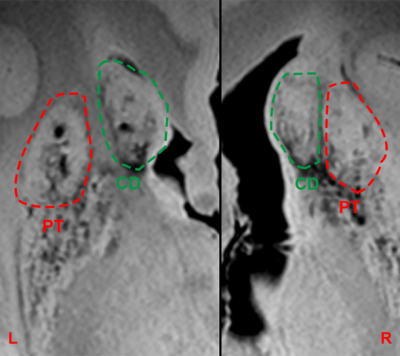
Figure 3: Susceptibility weighted imaging sequence (SWI) with a resolution of 0,15mm x 0,15mm x 0,5mm clearly identifies grafted tissue (dashed line) in the degenerated and iron rich striatum. The right grafts show a partial overgrowth. CD=caudate nucleus, PT=putamen.
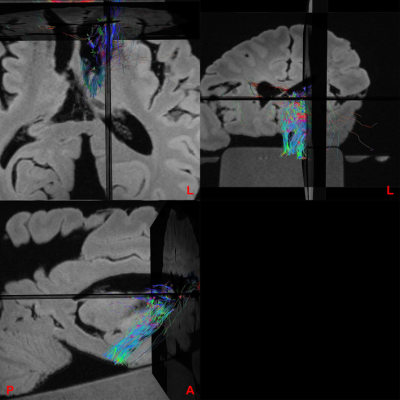
Figure 4: Three-plane overlay image between a SWI sequence and tracts derived from a diffusion tensor imaging (DTI) sequence with the left caudate nucleus graft serving as seed region. We found abundant fibers connecting mainly in direction of the brainstem, but also other regions of the brain, even contralateral.