1526
Investigation of subcortical brain structures in patients with Parkinson's disease using a quantitative susceptibility mapping atlas1School of Information Science and Technology, ShanghaiTech University, Shanghai, China, 2PET Center, Huashan Hospital, Fudan University, Shanghai, China, 3Department of Radiology, Ruijin Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, China, 4Institute for Medical Imaging Technology, School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China
Synopsis
A limited number of atlases have been constructed using quantitative susceptibility mapping (QSM) images from subjects with Parkinson’s disease (PD), and given disease-specific subcortical structures. In this work, we generated three standard-space templates i.e. the hybrid, QSM and T1w atlas, which kept good image quality to observe brain white, gray matter, and deep-brain nuclei. Based on the atlases, we achieved the manual annotation of a few brain subcortical structures, e.g. globus pallidus, substantia nigra, subthalamic nucleus and thalamus. The results gave the position and shape of subcortical nuclei which could be meaningful for the research and surgical treatment of PD.
Introduction
Human brain atlases are important for the research and treatment of Parkinson’s disease (PD), serving as references to identify anatomical structures 1. However, there are few atlases showing disease-specific subcortical structures from subjects with PD, and most of them are based on T1- and T2-weighted (T1w & T2w) images 2. In this work, we first constructed a hybrid human brain atlas using fused quantitative susceptibility mapping (QSM) and T1w images from 87 subjects with PD. It offered not only good contrast between cortical white and gray matter from T1, but also clear observation for iron-rich deep-brain nuclei from QSM. Moreover, we generated two individual QSM and T1w atlases by applying the same deformation fields on the original images respectively. Besides, we manually segmented 10 subcortical structures that are highly related to PD pathology on the atlases, including putamen (Pu), caudate nucleus (Ca), internal and external globus pallidus (GPi & GPe), red nucleus (RN), pars reticulata and pars compacta of substantia nigra (SNr & SNc), subthalamic nucleus (STN), habenular nuclei (HN) and thalamus (THAL). The thalamus was further segmented into 4 sub-regions, i.e. the anterior nuclei, the median nuclei, the lateral nuclei and the pulvinar.Methods
87 subjects (56.9±10.0 years old) were diagnosed with idiopathic PD according to the clinical diagnostic criteria of the UK Parkinson Disease Society Brain Bank 3. MRI scanning was performed at Rui Jin Hospital (Shanghai, China), using a 3.0 T MR system (Signa HDxt; GE Healthcare, Milwaukee, WI). Conventional T1w images with 1 mm isotropic resolution were acquired. A three-dimensional multi-echo GRE sequence was utilized to obtain T2*w images: (1) TR/TE1/spacing=59.3/2.7/2.9 ms, flip angle=12°, resolution 0.86×0.86×1.0 mm3; (2) TR/TE1/spacing=54.6/5.468/6.408 ms, flip angle=20°, resolution 0.47×0.47×2.0 mm3. All the images were resampled to the same resolution of 1×1×1 mm3 through operations in k-space. The raw phase was unwrapped using Laplacian-based phase unwrapping and the normalized background phase was removed by V-SHARP using the frequency shift. The susceptibility maps were determined by STAR-QSM algorithm to obtain the QSM images 4. All the programs were written by MATLAB R2011b (Mathworks, Natick, MA).The skull was removed from the T1w images using FSL BET, and then white and gray matter were segmented using FSL FAST. After this, the T1w images were normalized to the intensity range [0, 255] and co-registered to the corresponding magnitude images using FSL FLIRT. The hybrid images were calculated by fusing QSM and T1w images depending on the formula Hybrid=T1w-μ*QSM, where μ=400. The hybrid brain atlas was generated from all the hybrid images based on a group-wise registration method achieved by Advanced Normalization Tools (ANTs) 5. Meanwhile, we recorded the deformation fields and applied them on the original QSM and T1w images, respectively, to obtain two individual QSM and T1w atlases. Finally, we manually segmented 10 subcortical structures as described above using ITK-SNAP.
Results
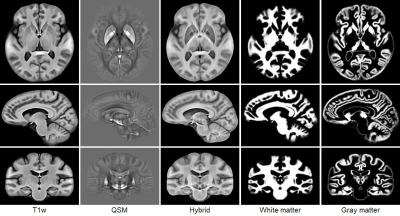
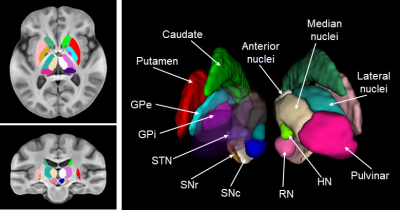
Figure 1 illustrates representative sections in the different views of the T1w, QSM and hybrid atlases, and the probabilistic segmentations of white and gray matter. Both the T1w and the hybrid atlases provide good performance of cortical contrast. Moreover, due to the fusion of information from the QSM images, the sections of the hybrid atlas provide better contrast for the subcortical structures, e.g. thalamus in the axial view, red nucleus and substantia nigra in the sagittal view. Figure 2 shows the manual annotations of subcortical brain nuclei overlaid on the sections of the hybrid atlas, and the 3D rendering is shown on the right. All the 10 labeled subcortical structures are displayed in the rendering image. It should be noted that thalamus is given by 4 sub-regions.Discussion and conclusion
The work constructed Parkinson Disease specific human brain atlases, which revealed the advantage of using QSM images to observe subcortical nuclei. The QSM template provided the feasibility of segmenting some structures into specific sub-regions. For instance, GP is labeled as internus and externus while SN as pars reticulata and pars compacta. In total, 10 subcortical structures were manually annotated based on the atlases. The position and shape of segmented subcortical structures can be helpful for the research and surgical treatment of PD, e.g. STN could be used to localize deep brain stimulation electrodes. Also, the atlases can also be warped into standard space to assist in studying human brain anatomy in neuroscience. Our perspectives concern the statistical analysis of magnetic susceptibility values in subcortical nuclei and combine with PET imaging of PD patients, to check the association and evaluate the ability of assisting the diagnosis..Acknowledgements
No acknowledgement found.References
1. Zhang Y, Wei H, Cronin MJ, et al. Longitudinal atlas for normative human brain development and aging over the lifespan using quantitative susceptibility mapping. NeuroImage. 2018;171;176-189.
2. Pauli W M, Nili A N, Tyszka J M, A high-resolution probabilistic in vivo atlas of human subcortical brain nuclei. Scientific Data. 2018;5:180063.
3. Hughes A J, Daniel S E, Kilford L, et al. Accuracy of clinical diagnosis of idiopathic Parkinson's disease: a clinico-pathological study of 100 cases. Journal of Neurology, Neurosurgery & Psychiatry. 1992;55(3):181-4.
4. Wei H, Dibb R, Zhou Y, et al. Streaking artifact reduction for quantitative susceptibility mapping of sources with large dynamic range. NMR in Biomedicine. 2015; 10:1294-303.
5. Avants Brian B, Yushkevich P, Pluta J, et al. The optimal template effect in hippocampus studies of diseased populations. Neuroimage. 2010;49(3):2457-2466.
Figures