1466
A preliminary study of geometric deep learning in brain morphometry: with application to Alzheimer’s disease
Huaiqiang Sun1, Lin Zou1, Haoyang Xing1, Min Wu1, Lei Chen2, Qing Li3, and Qiyong Gong1
1Huaxi MR Research Center (HMRRC), West China Hospital of Sichuan University, Chengdu, China, 2Department of Neurology, West China Hospital of Sichuan University, Chengdu, China, 3MR Collaborations, Siemens Healthcare Ltd., Shanghai, China
1Huaxi MR Research Center (HMRRC), West China Hospital of Sichuan University, Chengdu, China, 2Department of Neurology, West China Hospital of Sichuan University, Chengdu, China, 3MR Collaborations, Siemens Healthcare Ltd., Shanghai, China
Synopsis
A preliminary study that apply geometric deep learning to brain morphometry analysis of Alzheimer’s disease, the results show promising prediction accuracy suggesting geometric deep learning opens new prespectives for brain morphometry analysis of neurological and psychiatric disorders.
Introduction
Brain morphometry plays a fundamental role in neuroimaging research. Voxel-based morphometry (VBM) is currently the most widely used technology for neurological and psychiatric disorders. However, VBM aims to identify group level difference in the local composition of brain tissue, but hard to provide individual predictions1. The most difficult step for morphometry-based prediction is to extract quantitative features that describe the shape and also with appropriate dimensions. In recent proposed deep learning technique, the deep neural network learns useful representations and features automatically, directly from the raw data, bypassing this manual and difficult step. Compared with volume data, polygonal meshes provide a more efficient representation for brain morphometry, as mesh explicitly capture both shape surface and topology without been interfered by density variation. Research on deep learning has mainly focused so far on data defined on Euclidean domains. However, mesh data defined on non-Euclidean domains. The adoption of deep learning in these domain has been lagging behind until geometric deep learning was very recently proposed, which deals in this sense with the extension of deep learning techniques to graph/manifold structured data2. This preliminary study aims to apply geometric deep learning to classify brain meshes from patients with Alzheimer’s disease (AD) and matched controls.Methods
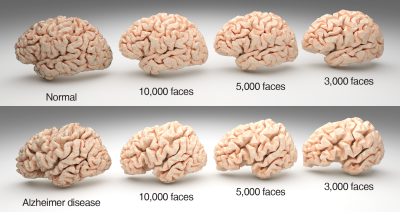
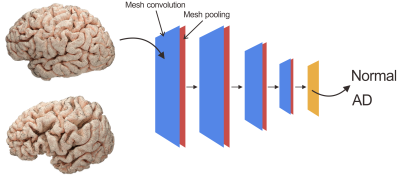
78 clinical confirmed AD patients (42 male, 36 female, aged 54 to 86 years, and mean age 71.2 years) were included in this study. 82 demographically-matched controls were recruited from the local community via poster advertisements. High resolution T1 weighted anatomical images were acquired on a clinical 3T scanner with a 12-channel phase array head coil using a MPRAGE sequence (TR=2400ms, TE=2ms, TI=1000ms, Flip angle=8, 1m isotropic resolution). T1 weighted anatomical images were processed with Freesurfer's recon-all processing pipeline. Both left and right pial surfaces were convert to triangular meshes. The raw meshes were corrected for topological defects and further simplified to number of faces of 10000, 5000 and 3000 using Blender 2.8 software (www.blender.org) (Fig. 1). 80% of subjects were randomly selected from each group to form the training set (62 ADs, 66 controls), and rest were assigned to test set (16 ADs, 16 controls). The simplified meshes in training set were send to an open source geometric deep learning framework named MeshCNN (ranahanocka.github.io/MeshCNN/)3. A schematic illustration of the deep neural network architecture is given in Fig. 2. The architecture contains 4 repeated blocks of an edge convolutional layer and an edge pooling layer.Results
For geometric deep neural networks trained on 10000, 5000 and 3000 faces of pial surface meshes of left hemisphere, the prediction accuracies on test set were 90.63%, 93.75% and 90.63%, respectively. The sensitivity for AD of three mesh simplification is 87.5%, 93.75% and 87.5%, the specificity is 93.75% in all three configurations. For neural networks trained on right hemisphere, the prediction accuracies on test set were 87.50%, 90.63% and 90.63%, respectively. The sensitivity for AD of three mesh simplification is 81.25%, 87.5% and 87.5%, the specificity is also 93.75% in all three configurations.Discussion
In this preliminary study, we utilize the unique properties of the mesh data for a direct classification of 3D brain shapes using geometric deep learning technique implemented in MeshCNN, a convolutional neural network designed specifically for triangular meshes, to identify AD patients from healthy controls. Analogous to classic CNNs, MeshCNN combines specialized convolution and pooling layers that operate on the mesh edges, by leveraging their intrinsic geodesic connections. According to the hold-out test, the trained network achieved satisfied prediction accuracy on test data. Due to the limitation of memory and the number of samples, we have to simplify the input model. However, the model simplification does not reduce the prediction accuracy of the network, probably due to the deep learning network has successfully captured the specific pattern of cortical atrophy caused by the disease. Based on our preliminary result, the geometric deep learning technique showed promising potential in the field of brain morphometry, especially for morphometry-based individual prediction. Moreover, the network architecture and the demand for sample size should be further investigated.Acknowledgements
No acknowledgement found.References
1. Ashburner J, Friston KJ. Voxel-Based Morphometry—The Methods. Neuroimage 2000;11:805–821
2. Bronstein MM, Bruna J, LeCun Y, Szlam A, Vandergheynst P. Geometric Deep Learning: Going beyond Euclidean data. IEEE Signal Process. Mag. 2017;34:18–42
3. Hanocka R, Hertz A, Fish N, Giryes R, Fleishman S, Cohen-Or D. MeshCNN. ACM Trans. Graph. 2019;38:1–12