1373
Deep learning based automated white matter lesion detection for MS and NMOSD: a retrospective multicenter study1Philips Healthcare, Suzhou, China, 2Beijing Tiantan Hospital, Capital Medical University, Beijing, China
Synopsis
Multiple sclerosis (MS) and neuromyelitis optical spectrum disorders (NMOSD) are common demyelination diseases in central neural system. With increasingly used MR examinations in clinical practice, the detection and delineation of white matter lesions is helpful to the calculation of total lesion volume, which is of great significance to clinical treatment planning. However, manual delineation is time-consuming to the clinicians and may lead to poor repeatability. In this work, we employed deep learning method to establish a tool for WM lesion delineation of MS and NMOSD routine MRI data from multiple centers.
Introduction
MRI has been widely used to detect the white matter lesions and it is important to accurately delineate the lesion and measure the lesion volumes for treatment planning. However, manual delineation by radiologists or clinicians is time-consuming and the intra- and inter-rater reproducibility is not guaranteed due to the intensive workload and the different perspectives from raters. This study aims to establish a deep learning (DL) based automated white matter lesion detection and delineation tool to assist the WM lesion identification work for radiologists, which could be applied to MR images from different clinical centers with MS or NMOSD lesions and with wide range of venders, image resolutions and slice thickness.Methods
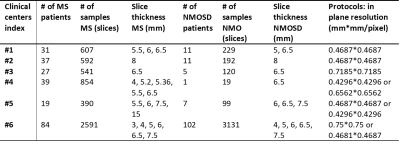
The MS and NMOSD data were collected from 6 clinical centers (Table 1). The clinical routine T2 FLAIR were acquired on 3T scanners from multiple vendors (Philips, GE and Siemens). Each patient data contained 17-30 slices to cover the whole brain area. A total of 237 MS patients and 137 NMOSD patients were included in this multi-center study. Manual labeling of the white matter lesions were performed by experienced radiologists with cross-check. Original data with in-plane resolution ranging from 0.4296×0.4296mm2/px to 0.75×0.75 mm2/px, and slice thickness ranging from 3mm to 15mm were resampled to matrix of 256×256 before network training. 80% of the patient data were arranged as training data from each clinical center in a balanced way. The rest were used for testing.We employed four DL network strategies for the neural networks: FCN (fully convoluted network 1) with upsample stride 16, 8 and 4 predictions back to pixels in a single step; and the Unet 2 strategy. The transferred learning was used, with implemented initial values from the Image-Net. For all the four networks, the encoder was used as VGG-19 3, and the loss function as the mean cross entropy, with mini-batch based stochastic gradient descent 4 and Adam optimizer 5. The learning rate was set to 3e-5. The network was implemented through Tensorflow, with GPU 1.4GHz, 1080Ti.
To quantitatively evaluate the performance of the networks, we calculated the prediction accuracy, which was defined as Accuracy=(TP+TN)/N, where the TP (true positive) was the lesion detection cases with Dice ratio larger than 0.5; TN (true negative) was the healthy slices with no lesion detected. The accuracy was calculated for the prediction from each clinical center.
Results
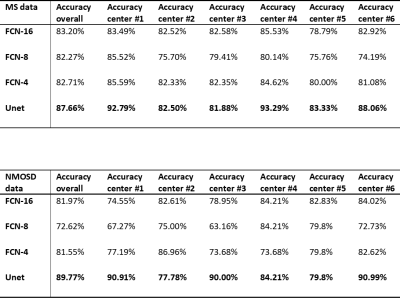
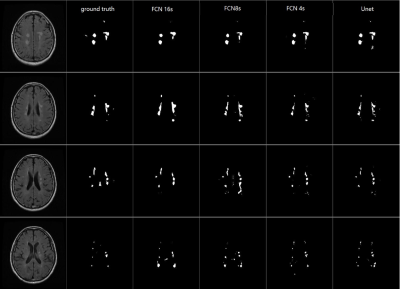
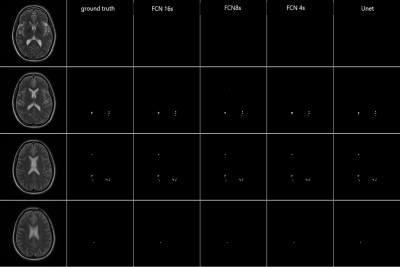
Figure 1 demonstrated the lesion detection from 4 DL network strategies. For the consecutive slices with MS lesions manually delineated, all the 4 networks could segment the white matter lesions consistently with the ground truth. The WM lesion for NMOSD case was shown in Figure 2. The small sized WM lesion in NMOSD consecutive slices were delineated consistently with the ground truth. Table 2 quantified the lesion detection accuracy of the 4 strategies for each disease type and from each data source center. The Unet achieved overall accuracy of 87.66% for MS and 89.77% for NMOSD, superior to the other 3 strategies, and achieved superior accuracies in the dataset from most centers. The accuracy on the whole testing set reached 88.64% with Unet strategy. The processing time for WM lesion delineation on one patient (with 30 slices) was within 10s.Discussion and Conclusion
We have established an automated white matter lesion detection and delineation tool using DL network. Through the comparison of 4 DL network strategies, the Unet outperformed with regard to the detection accuracy and it can be applied to a wide spectrum of MRI data in both MS and NMOSD data. Future work includes refining the DL network and improving the practical workflow.Acknowledgements
No acknowledgement found.References
1. Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. Proc IEEE Conf CVPR. 2015: 3431-3440.
2. Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. Int. Conf. Med. image computing and computer-assisted intervention. Springer, Cham, 2015: 234-241.
3. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556, 2014.
4. H. Robinds and S. Monro. A stochastic approximation method. Annals of Mathematical Statistics, 1951; 22: 400–407.
5. Adam Kingma, D. P., Ba, J. L.. Adam. A Method for Stochastic Optimization. Int. Conf. Learning Representations, 2015: 1–13.
Figures