1364
A cross-species link between deficient synaptic pruning and functional hyper-connectivity in autism1Functional Neuroimaging Laboratory, Istituto Italiano di Tecnologia, Rovereto, Italy, 2Biology Department, University of Pisa, Pisa, Italy, 3Neuromodulation of Cortical and Subcortical Circuits Laboratory, Istituto Italiano di Tecnologia, Genova, Italy, 4Department of Psychology, University of Cyprus, Nicosia, Cyprus, 5Laboratory for Autism and Neurodevelopmental Disorders, Istituto Italiano di Tecnologia, Rovereto, Italy, 6Center for Mind and Brain Sciences, University of Trento, Rovereto, Italy, 7Stanford University, Stanford, CA, United States
Synopsis
Altered brain functional connectivity is a hallmark finding in autism but the neural basis of this phenomenon remains unclear. We show that a mouse line reconstituting synaptic pruning deficits observed in postmortem autistic brains exhibits widespread functional hyper-connectivity, and that pharmacological normalization of synaptic aberrancies completely rescues behavioral and functional connectivity deficits. We also show that a similar connectivity fingerprint can be isolated in human rsfMRI scans of people with autism, and linked to overexpression of genes related to this dysfunctional pathway. Our results reveal a possible mechanistic link between deficient synaptic pruning and functional hyper-connectivity in autism.
Introduction
Post-mortem examinations have revealed an excess of excitatory synapses in the brain of children with autism spectrum disorders (ASD) [1, 2]. Recent investigations have linked this trait to hyper-activity of the mTOR pathway, resulting in synaptic pruning deficits [1]. ASD is also characterized by alterations in brain functional connectivity as measured with resting state functional MRI (rsfMRI) [3, 4]. These observations raise the question of whether and how mTOR-related deficient pruning affects rsfMRI dysconnectivity observed in ASD. Here, we mapped dendritic synaptic density, rsfMRI connectivity [5, 6] and social behavior in tuberous sclerosis 2 deficient (Tsc2+/-) mice, a mouse line that mechanistically reconstitutes the mTOR-dependent dendritic spine surplus observed in post mortem ASD brain samples [1]. A separate cohort of animals was subjected to a daily treatment with the mTOR inhibitor rapamycin with the aim to rescue the synaptic surplus, and corroborate a mechanistic link between synaptic traits and brain-wide connectivity. We finally sought to translate these findings across species, by isolating an analogous connectivity signature in publicly available rsfMRI scans of children with ASD (ABIDE-I) [7].Methods
Mouse studies. All experiments were carried out in accordance with Italian regulations governing animal welfare and protection (DL 26/2014). We used a cross-sectional treatment protocol with four cohorts of n=20 animals each: control Tsc2+/+ mice treated with rapamycin; control Tsc2+/+ mice treated with vehicle; mutant Tsc2+/- mice treated with rapamycin; mutant Tsc2+/- mice treated with vehicle. After daily treatment during pre-pubertal phase [1], Tsc2+/- and control mice underwent rsfMRI mapping at 7 tesla under halothane sedation [8] using a single-shot EPI sequence (TR/TE = 1200/15 ms, flip angle 30°, FOV 2.3 × 2.3 cm, matrix 100 × 100, 18 coronal slices, slice thickness 0.60 mm, 1920 volumes). Data were preprocessed as previously described [9] and inter-group differences in rsfMRI connectivity were mapped using weighted degree centrality and seed-based analysis [5, 10]. Post-mortem dendritic spine density was measured as in [1]. Human rsfMRI studies. We used weighted degree centrality and seed based analysis to map rsfMRI connectivity in pre-pubertal (age range 7-13) children with ASD (n=163) and typically developing controls (n=168) from eight sites of the ABIDE-I collection. Gene enrichment analyses were carried out as described in [11].Results and discussion
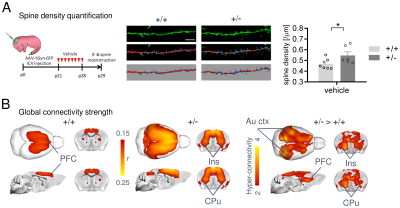
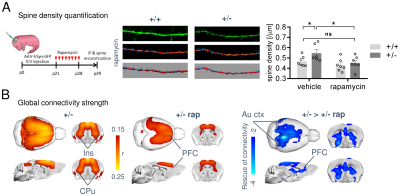
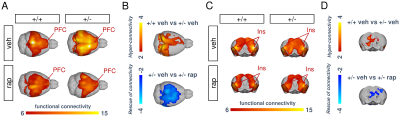
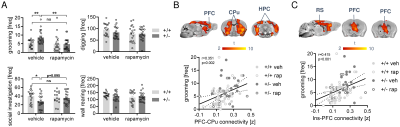
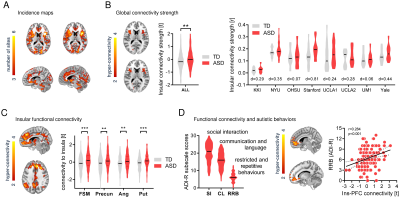
In keeping with previous findings, we observed increased spine density in juvenile Tsc2+/- mice (p < 0.05, Figure 1A), an effect associated with largely altered connectivity, involving long-range rsfMRI over-synchronization of integrative neocortical regions and the basal ganglia (Figure 1B). Importantly, developmental treatment with the mTOR-inhibitor rapamycin completely rescued synaptic density (p < 0.05, Figure 2A) and normalized rsfMRI hyper-connectivity (t > 2, p < 0.01 FWE cluster corrected, Figure 2B) in Tsc2+/- mice, hence corroborating a mechanistic link between aberrant mTOR activity and functional over-connectivity. This effect was especially prominent in the mouse default mode (Figure 3A-B) and salience networks (Figure 3C-D). Developmental treatment with rapamycin also rescued autism-like behavioral deficits in Tsc2 mutant mice (Fig. 4). Notably, voxel-wise correlational mapping revealed that functional hyper-synchronization of insular and striato-prefrontal regions was predictive of motor stereotypies in these mice (Figure 4). Collectively, these results establish a causal link between mTOR-related pruning deficits, fronto-insular hyperconnectivity and ASD-relevant motor stereotypies.Given the prominent implication of mTOR pathway dysfunction for human ASD [1], we hypothesized that a similar hyper-connected phenotype should be identifiable in clinical ASD populations. In keeping with this notion, rsfMRI connectivity mapping in pre-pubertal children with ASD from ABIDE revealed foci of increased long-range connectivity in the anterior insula, basal ganglia and prefrontal cortices of affected participants (Figure 5A-B). Furthermore, insular-prefrontal over-connectivity was significantly associated with repetitive behaviors (Figure 5C-D), hence reconstituting both the connectional and behavioral phenotype observed in Tsc2+/- mice. By coupling gene decoding and enrichment analysis, we finally found that TSC2/mTOR-network genes appear to be expressed in a similar topological pattern as the identified ASD hyper-connectivity phenotype, suggesting a putative mechanistic link between the observed hyper-connectivity and mTOR-related aberrant activity in human ASD.
Conclusion
Taken together, our results establish a cross-species mechanistic link between mTOR-dependent deficient synaptic pruning and rsfMRI over-connectivity in ASD.Acknowledgements
This work was supported by Simons Foundation Grants (SFARI 400101) to A. Gozzi. A. Gozzi was also supported by Brain and Behavior Foundation 2017 (NARSAD - National Alliance for Research on Schizophrenia and Depression) and the European Research Council (ERC - DISCONN, GA802371). M. Pagani was supported by European Union's Horizon 2020 research and innovation programme (Marie Sklodowska-Curie Global Fellowship – CANSAS, GA845065).References
1. Tang, G., et al., Loss of mTOR-Dependent Macroautophagy Causes Autistic-like Synaptic Pruning Deficits. Neuron, 2014. 83(5): p. 1131-1143.
2. Hutsler, J.J. and H. Zhang, Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain research, 2010. 1309: p. 83-94.
3. Nomi, J.S. and L.Q. Uddin, Developmental changes in large-scale network connectivity in autism. NeuroImage: Clinical, 2015. 7: p. 732-741.
4. Uddin, L.Q., K. Supekar, and C.J. Lynch, Salience network-based classification and prediction of symptom severity in children with autism. JAMA Psychiatry, 2013. 70(8): p. 869-879.
5. Pagani, M., et al., Deletion of autism risk gene Shank3 disrupts prefrontal connectivity. The Journal of Neuroscience, 2019: p. 2529-18.
6. Sforazzini, F., et al., Distributed BOLD and CBV-weighted resting-state networks in the mouse brain. Neuroimage, 2014. 87: p. 403-15.
7. Di Martino, A., et al., The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol Psychiatry, 2014. 19: p. 659-667.
8. Bertero, A., et al., Autism-associated 16p11.2 microdeletion impairs prefrontal functional connectivity in mouse and human BRAIN, 2018. 141(7): p. 2055–2065.
9. Suetterlin, P., et al., Altered neocortical gene expression, brain overgrowth and functional over-connectivity in Chd8 haploinsufficient mice. Cerebral Cortex, 2018. 28(6): p. 2192-2206.
10. Michetti, C., et al., The knockout of Synapsin II in mice impairs social behavior and functional connectivity generating an ASD-like phenotype. Cerebral Cortex, 2017. 27(10): p. 5014-5023.
11. Gorgolewski, K.J., et al., NeuroVault. org: a web-based repository for collecting and sharing unthresholded statistical maps of the human brain. Frontiers in neuroinformatics, 2015. 9: p. 8.
Figures