1301
Accurate Brain Extraction Using 3D U-Net with Encoded Spatial Information1Brain and Mind Centre, the University of Sydney, Sydney, Australia, 2Sydney Neuroimaging Analysis Centre, Sydney, Australia, 3Sydney Medical School, the University of Sydney, Sydney, Australia, 4I-MED Radiology Network, Sydney, Australia, 5Sydney Imaging and School of Biomedical Engineering, the University of Sydney, Sydney, Australia, 6School of Computer Science, the University of Sydney, Sydney, Australia
Synopsis
Brain extraction from 3D MRI datasets using existing 3D U-Net convolutional neural networks suffers from limited accuracy. Our proposed method overcame this challenge by combining a 3D U-Net with voxel-wise spatial information. The model was trained with 1,615 T1 volumes and tested on another 601 T1 volumes, both with expertly segmented labels. Results indicated that our method significantly improved the accuracy of brain extraction over a conventional 3D U-Net. The trained model extracts the brain from a T1 volume in ~2 minutes and has been deployed for routine image analyses at the Sydney Neuroimaging Analysis Centre.
Introduction
Brain extraction is a fundamental operation for brain tissue analysis. Currently, software packages such as FSL’s Brain Extraction Tool (BET)1 followed by manual QA and expert ‘cleaning’ are required to complete this task. A skilled neuroimaging analyst needs approximately 30 minutes to extract one accurate brain mask based on BET, which is tedious and time-consuming. Recently, deep convolutional neural networks including 3D U-Net2-4 have been proposed for brain segmentation. However, these methods ignore spatial information and have unsatisfactory accuracy. We overcame this challenge by encoding spatial information together with voxel values to train the network.Methods
Brain extraction was treated as a binary segmentation problem. For each voxel within the head volume, one of the two class tags (brain tissue or non-brain tissue) was assigned.(1) Brain extraction pipeline
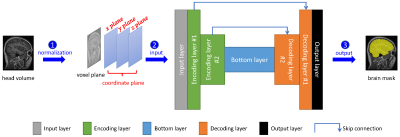
A pipeline with three steps for automatic brain extraction was designed (Figure 1). Firstly, using FSL,5 the native brain volume was registered into the MNI152 brain space with a unified normalized coordinate system. Each voxel encodes four values: voxel intensity, x coordinate, y coordinate and z coordinate. Secondly, we performed voxel normalization for each brain volume.6 The normalized voxel and coordinate values were fed into the 3D U-Net deep convolutional neural network. Finally, the brain mask generated by the network was transformed with FSL from MNI152 brain space back into the native data space.
(2) Coordinate alignment
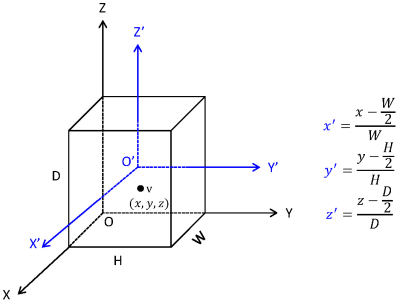
We developed a scheme to generate and normalize coordinate values of each voxel within head volume (Figure 2). Firstly, we established a coordinate system named OXYZ. Within the system, for each voxel v within a head volume with size DxWxH, we generated its coordinate values as (x, y, z). Coordinate values were then aligned into a new coordinate system named O’X’Y’Z’, yielding the aligned coordinate values, i.e. (x’, y’, z’).
(3) Training and testing
We adopted the patch extraction strategy,3,4 in which a cubic (patch) was randomly extracted from each head volume, to train the network in each training epoch. This strategy ensures that a sufficient number of training samples are available to the network. We set the patch size as 32x32x32.
For testing, each head volume was segmented into overlapping patches. As per training, we set the size of each patch as 32x32x32 and the overlapping step between adjacent patches as 8.
Results
2,216 T1-weighted typical clinical-quality head volumes (669 healthy control and 1,547 MS patients) acquired across different scanner models and field strength with ~1 mm3 voxel size were used in this study. Among these volumes, 1,615 scans were randomly selected as the training set, and the remaining 601 scans were used as the test set.(1) Qualitative results
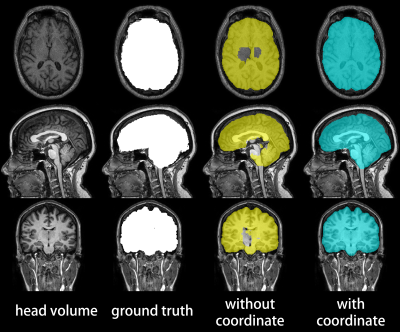
Figure 3 compares the gold-standard brain masks to the brain masks generated with and without coordinate information. Overt faults with brain masks generated without spatial information were corrected by our method, which generated masks judged as equivalent to or better than the gold-standard mask by a blinded examiner for all test volumes.
(2) Quantitative results
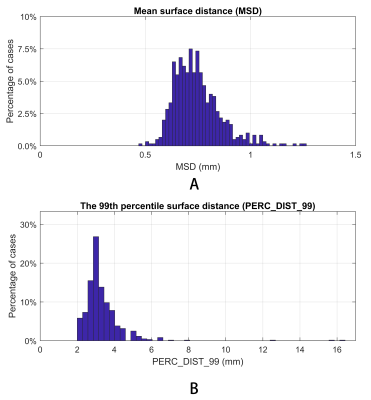
Figure 4 shows the Dice similarity coefficient (DC) (the higher the better), mean/root-mean-squared/95th percentile/99th percentile surface distance (the lower the better) between the model output and the ground truth. The proposed method achieves sub-voxel (<1 mm) average mean surface distance. The 95th and 99th percentile surface distances, which correspond to the most inaccurate sub-regions of each segmentation, were small in most cases, with a mean value of 2.0 mm and 3.3 mm, respectively. While cases with large 99th percentile surface distance exist, they are very rare. Figure 5 shows the distribution of the mean and 99th percentile surface distances. 96.5% of the cases had a mean surface distance <1 mm. The 99th percentile surface distance was 2-5 mm for the majority of the cases, with only 4.7% of the cases exhibiting >4 mm 99th percentile surface distance, indicating the model’s ability to prevent clinically significant segmentation failure.
Discussion
The capacity of deep networks to learn precise mapping between head voxels and brain masks is impeded by the data distribution and morphological variation among head scans. Encoding coordinate information together with voxel values provides spatial ‘hints’ to the network and improves the accuracy of brain extraction. With our proposed method, sub-millimeter mean surface distance (0.7 mm) was consistently achieved (0.1 mm standard deviation), indicating its ability to extract the brain with accuracy clinically equivalent to a human expert. The robustness of the model is indicated by 95.3% of the cases having <5 mm 99th percentile distance. The accuracy and robustness of the proposed method encourages its use in routine clinical practice.Conclusion
Our method enables accurate automatic brain extraction with typical clinical-quality data, which is an essential component of most clinical neuroimaging analyses. Compared with current brain extraction practice, which takes a skilled neuroimaging analyst ~30 minutes, our method extracts the brain automatically using only 2 minutes. This has the potential of enabling quantitative brain and tissue volume analyses in routine clinical practice. The proposed method has been used for routine research neuroimaging analyses at the Sydney Neuroimaging Analysis Centre and has shown improved accuracy and productivity compared to conventional manual workflow.Acknowledgements
This work was supported by an Australian Government Cooperative Research Centre Project (CRC-P) Grant.References
1. Smith SM. Fast robust automated brain extraction. Human Brain Mapping. 2002; 17(3): 143-155.
2. Ronneberger O, Fischer P, Brox T. U-Net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention. 2015; 234-241.
3. Wang ZY, Zou N, Shen DG, et al. Global deep learning methods for multimodality isointense infant brain image segmentation. arXiv preprint arXiv: 1812.04103, 2018.
4. Nie D, Wang L, Adeli E, et al. 3-D fully convolutional networks for multimodal isointense infant brain image segmentation. IEEE Transactions on Cybernetics. 2019; 49 (3): 1123-1136.
5. Smith SM, Jenkinson M, Woolrich MW, et al. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage, 2004; 23(S1):208-219.
6. TensorFlow. Per Image Standardization. https://www.tensorflow.org/api_docs/python/tf/image/per_image_standardization. Accessed October 14, 2019.
Figures