1295
Contrast-weighted SSIM loss function for deep learning-based undersampled MRI reconstruction1GE Research, Niskayuna, NY, United States, 2GE Healthcare, Menlo Park, CA, United States, 3GE Healthcare, Waukesha, WI, United States, 4Department of Radiology, Mayo Clinic College of Medicine, Rochester, MN, United States, 5Walter Reed National Military Medical Center, Bethesda, MD, United States
Synopsis
Deep learning-based undersampled MRI reconstructions can result in visible blurring, with loss of fine detail. We investigate here various structural similarity (SSIM) based loss functions for training a compressed-sensing unrolled iterative reconstruction, and their impact on reconstructed images. The conventional unweighted SSIM has been used both as a loss function, and, more generally, for assessing perceived image quality in various applications. Here we demonstrate that using an appropriately weighted SSIM for the loss function yields better reconstruction of small anatomical features compared to L1 and conventional SSIM loss functions, without introducing image artifacts.
Introduction
Recently, deep-learning techniques1-3 have been applied to MRI reconstruction from highly undersampled k-space data, attracting considerable interest due to their promise for improving image quality compared to PICS (parallel imaging and compressed sensing) methods. However, despite improved overall image quality, deep learning-based methods sometimes produce residual image blurring, with some loss of fine detail. The use of structural similarity (SSIM)4 as a loss function for network training was recently shown5 to produce sharper MR images than L2. And the combination of multi-scale SSIM and L1 outperformed other loss functions in super-resolution, denoising, and deblocking of natural images6. Here we explore the use of other variations of SSIM, including contrast-weighted SSIMs, as loss functions for undersampled MRI reconstruction.Methods
We use a DCI-Net (Densely Connected Iterative Network)3, an unrolled compressed-sensing iterative reconstruction consisting of multiple iterative blocks, each of which includes a data-consistency unit and a convolutional unit for regularization, with dense skip-layer connections among iterations. An example network was chosen with 20 iterations, 12 layers per iteration, 64 3x3 filters per layer, and 8 dense skip-layer connections per iteration.The SSIM function can be defined as4 $$\mathrm{SSIM}(x,y)=l(x,y)^{\alpha}c(x,y)^{\beta}s(x,y)^{\gamma}$$ for two image patches $$$x$$$ and $$$y$$$, where $$$l$$$, $$$c$$$ and $$$s$$$ are the luminance, contrast and structure comparison functions, and $$$\alpha$$$, $$$\beta$$$ and $$$\gamma$$$ are the weights. When $$$\alpha=\beta=\gamma=1$$$, this becomes the conventional unweighted SSIM. The comparison functions are given by $$l(x,y)=\frac{2\mu_x\mu_y+c_1}{\mu_x^2+\mu^2_y+c_1}$$ $$c(x,y)=\frac{2\sigma_x\sigma_y+c_2}{\sigma_x^2+\sigma_y^2+c_2}$$ $$s(x,y)=\frac{\sigma_{xy}+c_3}{\sigma_x+\sigma_y+c_3}$$ where $$$\mu_x$$$ and $$$\mu_y$$$ are the means of $$$x$$$ and $$$y$$$, $$$\sigma_x$$$ and $$$\sigma_y$$$ are the variances, $$$\sigma_{xy}$$$ is the covariance, $$$c_1=(k_1L)^2$$$, $$$c_2=(k_2L)^2$$$, and $$$c_3=c_2/2$$$ with $$$k_1=0.01$$$, $$$k_2=0.03$$$ and $$$L$$$ is the dynamic range. Given a network output image and a fully-sampled reference image, the SSIM is calculated for 11x11 image patches, centered around each pixel of the two images, and the mean of the SSIM over the entire image is used as the loss function. We have used different loss functions for training the network, including L1, the conventional unweighted SSIM4, and various weighted SSIM (wSSIM), where $$$\alpha$$$, $$$\beta$$$ and/or $$$\gamma$$$ can deviate from 1. As one example, a contrast-weighted grouping of $$$\alpha=0.3$$$, $$$\beta=1$$$, and $$$\gamma=0.3$$$ was chosen for closer analysis based on preliminary visual assessment of reconstructed images with respect to blurring, noise, and artifacts.
Fully-sampled T1 and T2-weighted fast-spin-echo MRI head datasets were acquired using a variety of 8 to 49-channel head arrays. The data were retrospectively down-sampled with a net undersampling factor of 3.7 and used for training and testing. In total, 3062 slices were collected, of which 1901 were used to train the networks, 151 for validation and 1010 for testing. The test data were acquired at the Mayo Clinic on their compact 3T scanner7. Mixtures of uniform and variable-density undersampling were used for training, with variable-density sampling used for testing. Complex coil-combined zero-filled reconstructions from the down-sampled data were used as the input to the network, and the fully sampled k-space data were reconstructed to form reference images.
Reconstructed images were quantitatively compared against image metrics relative to fully-sampled ground truth including normalized mean-squared error (nMSE), peak signal-to-noise ratio (PSNR), SSIM and wSSIM. Here we also compared with Autocalibrating Reconstruction for Cartesian imaging (ARC) – where k-space was regularly undersampled, and with a total-variation (TV) based compressed-sensing method8 that employed data-driven iterative soft thresholding.
Results
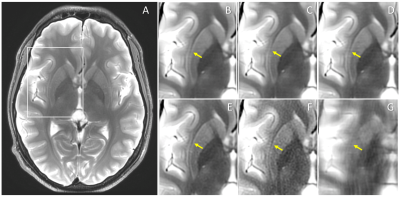
Figures 1 and 2 show representative examples of the test images, comparing the output of the DCI-Net trained using different loss functions. Here wSSIM resulted in better recovery of fine anatomical features than the other loss functions. Table 1 gives a comparison of the quantitative image metrics for different reconstruction methods, calculated from the test data set. The three networks outperformed TV and ARC in all metrics, with the two SSIM losses generating better SSIM metrics than the L1 loss.Discussion
It has been reported5,6 that L1 and SSIM loss functions and their variants and combinations can yield better image quality such as improved sharpness compared to conventional L2. In this study, the wSSIM loss function showed better fine-feature recovery than L1, or SSIM, at the cost of somewhat weaker image denoising. The wSSIM loss function encourages similarity in local spatial variance between the network output and the fully-sampled reference image, which appears to improve reconstruction of small anatomical features. The new wSSIM metric merits further investigation, including examining its variants and combination with other loss functions.Conclusion
The novel weighted SSIM loss function shows promise for reconstructing small anatomical features with better image quality compared to L1 and conventional SSIM loss functions, when applied to reconstructions from highly undersampled k-space data.Acknowledgements
References
1. Hammernik K, et al. Learning a variational network for reconstruction of accelerated MRI data. Magn Reson Med 2018;79(6):3055-3071.
2. Schlemper J, et al. A deep cascade of convolutional neural networks for dynamic MR image reconstruction. IEEE Trans Med Imaging 2018;37(2):491-503.
3. Malkiel I, et al. Densely connected iterative network for sparse MRI reconstruction. ISMRM 2018, p. 3363.
4. Wang Z, et al. Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process 2004;13(4):600-612.
5. Hammernik K, et al. L2 or not L2: Impact of loss function design for deep learning MRI reconstruction. ISMRM 2017, p. 687.
6. Zhao H, et al. Loss functions for image restoration with neural networks. IEEE Trans Comput Imaging 2016;3(1):47-57.
7. Foo TKF, et al. Lightweight, compact, and high-performance 3T MR system for imaging the brain and extremities. Magn Reson Med 2018;80(5):2232-2245.
8. Khare K, et al. Accelerated MR imaging using compressive sensing with no free parameters. Magn Reson Med 2012;68(5):1450-1457.
Figures