1286
Toward a site and scanner-generic deep learning model for reduced gadolinium dose in contrast-enhanced brain MRI1Subtle Medical Inc., Menlo Park, CA, United States
Synopsis
Gadolinium-based contrast agents (GBCAs) create unique image contrast to facilitate identification of various clinical findings. However, recent discovery of gadolinium deposition after contrast-enhanced MRI raises new safety concerns of GBCAs. Deep learning (DL) has recently been used to predict the contrast-enhanced images using only a fraction of the standard dose. However, challenges remain in generalizing the DL methods across different protocols/vendors/institutions. In this work, we propose comprehensive technical solutions to improve DL model robustness and obtain high quality low-dose contrast-enhanced MRI across multiple scanners and institutions.
Introduction
Gadolinium-based contrast agents (GBCAs) have been indispensable in MRI exams for investigating pathology and prognosis. Unfortunately, the identification of prolonged gadolinium deposition within the brain and body has raised safety concerns [1-4]. Recently, a deep learning (DL) framework was presented [5] that could reconstruct contrast-enhanced images while reducing GBCA dose levels by 10 times and maintaining non-inferior image quality. While promising, the study was limited to scans from a single institution and the images were independently processed as 2D slices. The generalizability of the DL method for diverse clinical settings is unclear. Furthermore, a robust DL solution should retain the multiplanar reformat (MPR) capability for 3D images to enable oblique visualizations. This work proposes comprehensive technical solutions to develop a generalized robust model addressing the aforementioned challenges.Methods
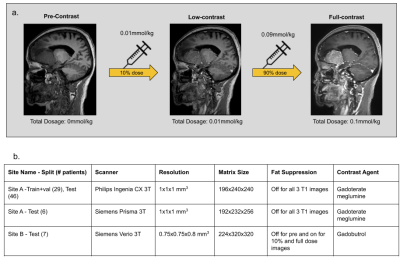
Data & PreprocessingWith IRB approval and informed consent, the scanning protocol shown in Fig.1a was implemented in two institutions. The institution-scanner distribution and variability of the train and test datasets are tabulated in Fig.1b. For each participant, three 3D T1-weighted images were obtained sequentially in one imaging session: pre-contrast, 10% dose(0.01mmol/kg), and full dose (0.1mmol/kg). Preprocessing steps involve signal normalization through histogram matching to account for differing receive gains between acquisitions, followed by affine co-registration between the three images[6]. The DL model was designed with the assumption that the contrast-enhanced signal between pre-contrast and low-dose images was non-linearly related to that of full-dose images.
DL Model Design
The following technical solutions were applied to achieve greater model robustness and generalizability.
(1) 2.5D Training
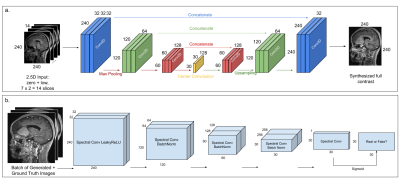
We used the U-Net architecture from [5] with seven adjacent pre and 10%-contrast image slices stacked across channels as the input (Fig.2a), to reduce streaking artifacts and to improve contrast in structures that span multiple slices. The DL model was trained with a combination of L1 and structural-similarity index (SSIM) losses, predicting the central full contrast slice.
(2) MPR Reconstruction
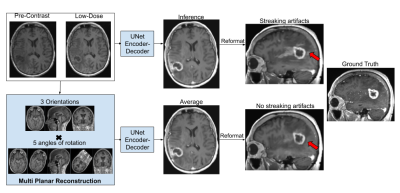
Although the 2.5D model reduced streaking artifacts in orthogonal planes (Fig.3), it was not sufficient to remove them in cases with dense tissue structures, as the 2.5D processing neglects the true volumetric nature of the acquisition. To facilitate learning complex 3D structures without producing MPR artifacts while maintaining memory efficiency of 2D processing, we rotated the 3D training volumes at 15 equispaced angles from 0-360deg before taking the respective slices for mini-batch training. During inference, we do a similar processing to produce 15 output volumes and average those to obtain the final result (Fig.3).
(3) Perceptual & Adversarial Losses
To restore texture and improve image resolution, we added perceptual loss from the VGG19 network [7,8] and introduced adversarial training [9], where a patch discriminator [10] was trained in parallel to the U-Net generator. Fig.2b shows the architecture of the patch discriminator, consisting of a series of spectral normalized [11] convolutional layers with LeakyReLU activations and predicts a 30x30-patch. The discriminator was trained with MSE loss, with gaussian noise added to the inputs to facilitate smooth convergence [12].
(4) Enhancement Mask
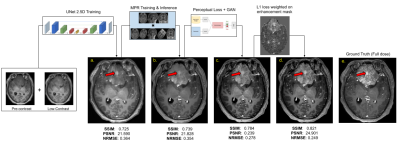
Although prior signal normalization reduces the relative scaling difference between scans, some enhancement-like differences still remain. To focus the learning to actual enhancement regions, we weighted the L1-loss with a continuous enhancement-mask computed from the difference of the skull-stripped [13] pre-contrast and 10%-contrast (Fig.4c&d) so that enhancement regions have larger impact on the prediction.
Problem Formulation
Considering the above improvements, the learning problem can be formulated as follows: $$G^* = argmin_G [\lambda_{GAN} L_{GAN}(G) + \lambda_{L1}L_{L1}(M_{enh} \cdot G) + \lambda_{SSIM}L_{SSIM}(G) + \lambda_{VGG} L_{VGG}(G)] $$where $$$ L_{GAN} = max_D L_{GAN}(G,D)$$$ (where G=U-Net-Generator and D=discriminator) and $$$M_{enh}$$$ is the smooth enhancement mask and the respective lambdas were chosen empirically on the validation set.
Experiments
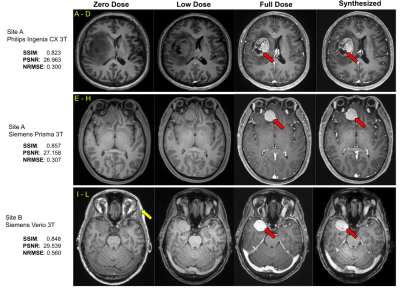
Using the 29 cases (24-train/5-validation) from Site-A Philips scanner, all models were trained on Nvidia-GeForce-RTX2080 GPUs for 100-epochs with Adam optimizer with an initial learning rate of 0.001. The model generalizability was tested on the 46 cases of the same site/scanner. The results were evaluated using PSNR, SSIM and RMSE, considering true full-dose as reference. Cross-vendor and cross-site generalizability were tested using the 6 cases from Site-A Siemens scanner and the 7 cases from Site-B Siemens scanner respectively.
Results & Discussion
As shown in Fig.4, different components in the DL model design progressively contributed to improved image quality. The average PSNR and SSIM on the test set for the original(proposed) model is 25.275(28.254) and 0.830(0.892) respectively. The best results were achieved with MPR and a combination of SSIM, perceptual, adversarial and enhancement-weighted L1 losses.Based on the qualitative and quantitative results, the addition of various technical enhancements has led to a significant improvement in model robustness and generalizability (Fig.5). Though MPR-processing increased the inference time, it provided a solution that feasibly fits on GPU memory. Here, we have demonstrated that a model trained with images from a single-site/single-scanner is able to generalize well on images from other scanners and sites. We will broaden the scope of the generalizability test and assess the clinical performance in future work.
Conclusion
We have proposed comprehensive technical solutions for low-dose contrast-enhanced MRI using DL. Initial studies validated the robustness and generalizability of the proposed method in diverse clinical settings, a significant step towards its wide clinical adoption.Acknowledgements
We would like to acknowledge the grant support of NIH R44EB027560.References
1. Brasch RC, Weinmann HJ, Wesbey GE. Contrast-enhanced NMR imaging: animal studies using gadolinium-DTPA complex. Am J Roentgenol 1984;142:625–630.
2. Carr DH, Brown J, Bydder GM, et al. Gadolinium-DTPA as a contrast agent in MRI: initial clinical experience in 20 patients. Am J Roentgenol 1984;143:215–224.
3. Silver NC, Good CD, Barker GJ, et al. Sensitivity of contrast enhanced MRI in multiple sclerosis. Effects of gadolinium dose, magnetization transfer contrast and delayed imaging. Brain 1997;120:1149–1161.
4. McFarland HF, Frank JA, Albert PS, et al. Using gadolinium-enhanced magnetic resonance imaging lesions to monitor disease activity in multiple sclerosis. Ann Neurol 1992;32:758–766.
5. E. Gong, J. M. Pauly, M. Wintermark, and G. Zaharchuk. Deep learning enables reduced gadolinium dose for contrast-enhanced brain MRI, Journal of Magnetic Resonance Imaging, vol. 48, no. 2, pp. 330–340, 2018.
6. Marstal K, Berendsen F, Staring M, Klein S. 2016. SimpleElastix: A user-friendly, multilingual library for medical image registration. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops. 134–142.
7. Karen Simonyan, Andrew Zisserman. Very Deep Convolutional Networks for Large-Scale Image Recognition. ICLR 2015
8. Justin Johnson, Alexandre Alahi, Li Fei-Fei. Perceptual Losses for Real-Time Style Transfer and Super-Resolution arXiv:1603.08155 2016
9. Ian J. Goodfellow, Jean Pouget-Abadie, Mehdi Mirza, Bing Xu et al. Generative Adversarial Nets. Neural Information Processing Systems (NeurIPS) proceedings 2014
10. Phillip Isola, Jun-Yan Zhu, Tinghui Zhou, Alexei A. Efros. Image-to-Image Translation with Conditional Adversarial Networks. CVPR 2017
11. Takeru Miyato, Toshiki Kataoka, Masanori Koyama, Yuichi Yoshida. Spectral Normalization for Generative Adversarial Networks. ICLR 2018
12. Simon Jenni and Paolo Favaro. On Stabilizing Generative Adversarial Training with Noise. CVPR 2019
13. Deepbrain - https://github.com/iitzco/deepbrain
Figures