1234
A magnetization transfer weighted anatomical reference allows laminar fMRI analysis in native functional image space1Section on Functional Imaging Methods, Laboratory of Brain and Cognition, NIMH, NIH, Bethesda, MD, United States, 2Functional MRI Core, NIMH, NIH, Bethesda, MD, United States, 3Advanced MRI Section, Laboratory of Functional and Molecular Imaging, NINDS, NIH, Bethesda, MD, United States, 4Maastricht Brain Imaging Center, Faculty of Psychology and Neuroscience, University of Maastricht, Maastricht, Netherlands
Synopsis
In most previous laminar fMRI studies, cortical layers are defined based on an anatomical image that is collected by a different acquisition technique and exhibits different geometric distortion compared to the functional images. We introduce to generate a magnetization transfer (MT) weighted anatomical reference, using identical acquisition design as fMRI measurement. Cortical surface and depth can be reconstructed directly from this MT-weighted anatomical EPI image and all laminar analysis can be performed in the native fMRI image space without the need for distortion correction and registration.
Introduction
Laminar fMRI allows discrimination of information flow in cortical circuits, as afferent and efferent connections terminate in different cortical layers1. In most previous studies, the definition of cortical depth is based on an anatomical reference image that is collected by a different acquisition sequence and exhibits different geometric distortion compared to the functional images. Here, we propose to generate the anatomical image with the fMRI acquisition technique by incorporating magnetization transfer (MT) weighted imaging. This allows sufficient gray-white matter contrast to perform all laminar fMRI analysis in the native EPI space, without the need for distortion correction and registration.Theory and Methods
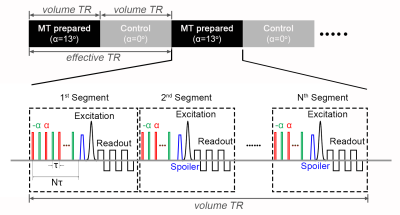
In the human brain, a relatively large fraction of macromolecular hydrogen protons (MP) (f~0.2-0.3) is found in white matter (WM), primarily due to its high content of myelin, while this number is smaller in gray matter (GM) (f~0.1)2. Through magnetization transfer (MT) with water hydrogen protons (WP), MPs can dramatically affect the MRI signal and thus different MP fractions will result in different MRI signal intensities. Here we applied a T2-selective pseudo-continuous pulse train to saturate the longitudinal magnetization of MPs based on their short T2 (<100us)3.The sequence is implemented to acquire images alternately between MT-prepared and CTRL conditions (Fig. 1). In MT-prepared condition, 3D-EPI readout-modules4 are interleaved with MT pulses to image the WP signal affected by MT. The MT preparation consists of a series of low flip-angle (α) pulses with alternating polarity. In CTRL condition, the preparation module is switched off by setting α=0°.
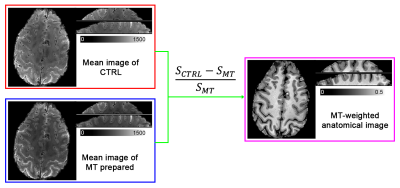
The MT-weighted anatomical image is generated as (SCTRL-SMT)/SMT, where SCTRL is the image signal in CTRL condition, and SMT is the image signal of MT-prepared condition (Fig. 2). This combination approach extracts the MT-saturated signal and removes the T2* contrast associated with the EPI readout.
Experiments
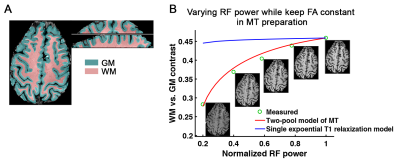
Five healthy volunteers (two females) participated this study. MT-3D-EPI was implemented on a Siemens MAGNETOM 7T scanner. Data was acquired with MT preparation parameters: pulse number (N) in 1st/later segment=250/36, flip angle (α)=13°, pulse interval (τ)=1.1ms, pulse duration=160us, and acquisition parameters: TE=24-28ms, excitation FA=25°, resolution=0.8mm isotropic, partial-Fourier of 7/8 (or 6/8 for whole brain) and GRAPPA 3. The 3D-EPI is segmented with the same number as slices and each segment takes about 100ms with its MT-preparation included. For the whole brain coverage, we acquire 96 slices with 8 slices oversampling. We average multiple measurements over 10 minutes to achieve sufficient image signal-to-noise ratio (SNR).To demonstrate that WM-GM contrast of MT-3D-EPI anatomical image is mainly contributed by MT effect rather than T1-weighting, we varied the RF power of preparation pulses while keeping their FAs constant. In this way, RF effect on the WP stays the same while the MP pool is saturated at different levels. The WM-GM contrast is evaluated as (SWM-SGM)/((SWM+SGM)/2), where SWM and SGM are the signal in WM and GM on the calculated anatomical image, respectively. The highest RF power allowed by the SAR limit was used for all other MT-weighted anatomical acquisition.
Results and discussion
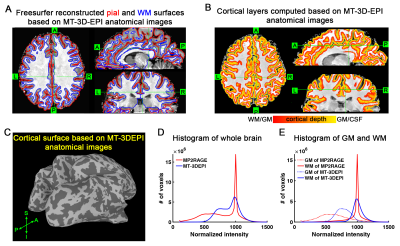
Fig. 3 shows WM-GM contrasts under different RF powers. The measured WM-GM contrast can be predicted well by the two-pool model of MT effect (Fig. 3B). Higher RF power results in more MP saturation which transfers to WP, and eventually leads to a larger WM-GM contrast due to their difference in MP fraction. On the contrary, when only considering the single exponential T1 relaxation of GM and WM, there is negligible variation of WM-GM contrast as FA is constant, which does not match experimental observation.Then we investigate the capability of this MT-weighted anatomical imaging for the whole brain coverage (cerebellum not included) and compared its WM-GM contrast with MP2RAGE images (Fig. 4). With this whole brain MT-weighted EPI images, we are able to run a freesurfer surface reconstruction (Fig. 4A and 4C) and then a cortical layer computation (Fig. 4B) automatically. Fig. 4E and 4F compares the histograms of MT-3D-EPI and MP2RAGE image intensities, across the whole brain, GM and WM regions. The GM and WM peaks are well separated in the red MP2RAGE curves, still good but less so in the blue MT-3D-EPI data. Nevertheless, the WM-GM contrast of MT-3D-EPI is sufficient to reconstruct the pial and WM surfaces automatically.
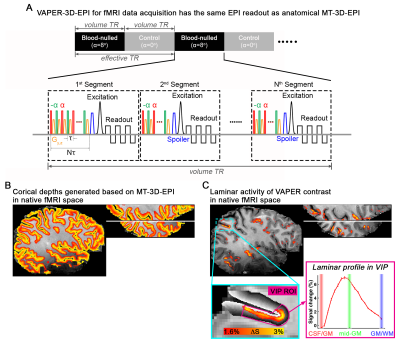
Laminar fMRI analysis can be achieved in native fMRI space when fMRI data and anatomical reference are acquired with an identical EPI readout. Here we acquire fMRI time series using an integrated vasodilation and perfusion contrast (VAPER)5. The functional VAPER sequence is designed identically to MT-3D-EPI, except that RF pulses in the preparation have relatively low FAs (8°) and a gradient pulse is added after each of them to make it most sensitive to intravascular signal changes (Fig. 5A). In the native fMRI space, cortical layers can be defined based on the anatomical images of MT-3D-EPI (Fig. 5B) and VAPER activity can be revealed in different laminar coordinates (Fig. 5C).
Conclusions
We introduced a MT-3D-EPI sequence approach to acquire an anatomical reference in the same space as fMRI data. It provides a sufficient WM-GM contrast (45% signal difference) which allows us to reconstruct cortical surface using automatic program and do all laminar analysis in native fMRI data space.Acknowledgements
This work was supported by the NIMH Intramural Research Program.References
1. Huber L, Handwerker D, Jangraw D, et al. High-Resolution CBV-fMRI Allows Mapping of Laminar Activity and Connectivity of Cortical Input and Output in Human M1. Neuron. 2017; 96: 1253-1263.
2. Van Gelderen P, Jiang X, Duyn J. Rapid measurement of brain macromolecular proton fraction with transient saturation transfer MRI. Magnetic resonance in medicine. 2017; 77: 2174-2185.
3. Wilhelm M, Ong H, Wehrli S, et al. Direct magnetic resonance detection of myelin and prospects for quantitative imaging of myelin density. Proceedings of the National Academy of Sciences of the United States of America. 2012; 109: 9605-9610.
4. Poser B, Koopmans P, Witzel T, et al. Three dimensional echo-planar imaging at 7 Tesla. Neuroimage. 2010; 51: 261-266.
5. Chai Y, Li L, Huber L, et al. A Novel Intravascular Contrast for Laminar Functional MRI. ISMRM. 2018.
Figures