1225
Highly Accelerated Sub-Millimeter Resolution 3D GRASE with Controlled T2 Blurring in T2-Weighted FMRI at 7T: Feasibility Study1University of California, Berkeley, Berkeley, CA, United States, 2Advanced MRI Technologies, Sebastopol, CA, United States
Synopsis
3D GRASE is used for cortical layer and columnar fMRI in the absence of signal confounds from draining veins. Its use has been limited by limited slice coverage with blurring. We developed highly accelerated 3D GRASE with controlled T2 blurring by combining compressed sensing with variable flip angles. Compared with current GRASE acquisitions, the proposed method demonstrates that 1) through-plane random encoding with VFA increases the slice coverage with a sharper point spread function, 2) reduced TE from in--plane random encoding provides a high SNR efficiency, and 3) the resulting image sharpness and SNR efficiency lead to increased BOLD activation.
Introduction
At ultra-high magnetic field, improved SNR makes it possible to acquire T2-weighted BOLD signals with high spatial specificity for columnar and layer imaging by minimizing draining vein effects1-3, in which BOLD effects originating from larger diameter draining veins can be significantly distant from the actual sites of neuronal activity. 3D gradient and spin echo imaging with inner-volume selection (GRASE) have been appealing options to achieve sub-millimeter resolution in fMRI, in which the required number of phase encoding steps is reduced at the same resolution by limiting the field-of-view4-6. In general, 3D GRASE has disadvantages in that 1) k-space filtering causes T2 blurring by limiting the number of slices and 2) a variable flip angle (VFA) scheme results in partial success with substantial SNR loss 7,8. To address these issues, we developed highly accelerated 3D GRASE with controlled T2 blurring through an optimal combination of compressed sensing9 and variable flip angles10,11. Numerical and experimental studies confirm three advantages of CS VFA GRASE: 1) through-plane random encoding with VFA increases slice number and narrows the point spread function (PSF), 2) reduced TE from in-plane random encoding provides a high SNR efficiency, and 3) the reduced blurring and higher tSNR result in higher BOLD activations.Methods
Sampling Design: Since an echo spacing is elongated to accommodate a large number of echoes, randomly undersampled k-space may cause significant signal discontinuity with a shuffled ordering across time as well as contrast changes with varying TE from undersampled EPI train, potentially yielding undesirable artifacts with temporal noises. To mitigate signal discontinuity across time, k-space is segmented for partition encoding by 1) dividing it into several frequency bands along kz direction according to a probability function and 2) randomly choosing one line within a single band. For phase encoding, the same number of ky lines before the center k-space ky=0 is acquired with random blips for a constant TE across time (Fig. 1).Variable Flip Angle: To control T2 blurring along the partition direction, the VFA regime, calculated using an inverse solution of Bloch equations based on a GM-specific prescribed signal evolution (T1=2000ms and T2=60ms), was used to achieve slow signal decay. The prescribed signal was described as: $$$ \mathrm{y_m =exp(-\frac{\beta(m-1)T_{ESP}}{ETL \times T_{ESP}})}$$$ where m is the number of refocusing pulses, ETL is the echo train length, and $$$\beta$$$ is a calibration factor that determines the signal decay rate.
Reconstruction: Since fMRI time series of images can be represented as a linear combination of a background tissue, which is slowly varying across time, and a dynamic BOLD signal, which is rapidly changing depending on stimulation, the reconstruction prior for each component needs to be correspondingly different. Assuming that the background signal lies in a low dimensional subspace while its residual is sparse in a transform domain, the undersampled fMRI data is reconstructed using low rank (for background tissue) and sparsity (for dynamic activation) priors known as k-t RPCA12-14.
Experiments: fMRI studies were conducted on a 7T whole body MR scanner (MAGNETOM 7T, Siemens Medical Solution, Erlangen, Germany) equipped with a 32-channel head coil. Institutional review board and informed consent was obtained for all subjects. All data were acquired using 1) Normal-, 2) VFA-, and 3) CS VFA-GRASE. In all experiments, the spatial and temporal resolutions were set to 0.8 mm (isotropic) and TR=3s, respectively, with 92 time frames, resulting in total duration of 4.6 minutes. Other imaging parameters are summarized in Table 1.
Stimulation Paradigm: Functional activation was assessed by performing a vertical meridian localizer stimulus, which alternated between 15s of visual stimulation at the horizontal meridian followed by 15s at the vertical. The cycle was repeated 9 times per scan with on additional 9s at the beginning of the scan to reach steady state.
Results
To investigate the signal behaviors of GM tissue with CFA and VFA schemes, the VFA and its signal evolution were estimated with varying β=-0.8, -1.5, and -4.2 for 24 and 36 slices (Fig. 2). In the CFA scheme, the signal intensity of GM decreases rapidly, thus yielding severe blurring in the coronal plane. On the other hand, the signal slope of the VFA gradually reduces with decreasing β, yielding the reduced blurring at the cost of mean tSNR. Figure 3 shows the simulated PSFs and activation maps. The activation demonstrates that the CFA introduces a BOLD smearing effect across neighboring regions, while the VFA improves spatial specificity at the cost of sensitivity and then gradually reduce the activation with decreasing β.To evaluate the performance of CS VFA GRASE against different GRASE methods, four sets of the visual cortex data were acquired. The proposed method describes the activation in the vicinity of GM with high sensitivity by minimizing a trade-off between sensitivity and specificity (Fig. 4).
Conclusion
We demonstrated that CS VFA GRASE has significant improvement over normal and VFA GRASE in terms of PSF, tSNR, and increased image volume in measuring functional activation. Unlike normal and VFA GRASE methods that must balance a tradeoff between the above measures, the proposed method is able to minimize these dependencies without an apparent loss of information.Acknowledgements
This work has been supported through NIBIB U01EB025162, NINDS R44NS084788, and NIMH R01MH111444, R44MH112210.References
1. Uludag K, Muller-Bierl B, Ugurbil K. An integrative model for neuronal activity-induced signal changes for gradient and spin echo functional imaging. Neuroimage. 2009;48:150165.
2. Yacoub E, Van De Moortele PF, Shmuel A, Ugurbil K. Signal and noise character-istics of Hahn SE and GE BOLD fMRI at 7 T in humans. Neuroimage. 2005; 24:738750.
3. Duong TQ, Yacoub E, Adriany G, Hu X, Ugurbil K, Kim SG. Micro-vascular BOLD contribution at 4 and 7T in the human brain: gradient-echo and spin-echo fMRI with suppression of blood effects. Magn Reson Med. 2003; 49: 10191027.
4. Feinberg DA, Hoenninger JC, Crooks LE, Kaufman L, Watts JC, Arakawa M. Innervolume MR imaging: technical concepts and their application. Radiology. 1985; 156:743747.
5. Feinberg DA, Harel N, Ramanna S, Ugurbil K, Yacoub E. Sub- millimeter single-shot 3D GRASE with inner volume selection for T2 weighted fMRI applications at7 Tesla. In Proceedings of the 16th Annual Meeting of ISMM, Toronto, Ontario,Canada, 2008. Abstract 2373.
6. Feinberg DA, Oshio K. GRASE (gradient- and spin-echo) MR imaging: a new fast clinical imaging technique. Radiology. 1991;181:597602.
7. Kemper VG, De Martino F, Vu AT, Poser BA, Feinberg DA, Goebel R, Yacoub E. Sub-millimeter T2 weighted fMRI at 7 T: comparison of 3D-GRASE and 2D SE-EPI.Front Neurosci. 2015;9:163.
8. Kemper VG, Martino FD, Yacoub E, Rainer G. Variable flip angle 3D-GRASE for high resolution fMRI at 7 tesla. Magn Reson Med. 2016; 76: 897–904.
9. Lustig M, Donoho D, Pauly JM. Sparse MRI: the application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007; 58: 11821195.
10. Mugler JP, III, Bao S, Mulkern RV, Guttmann CR, Robertson RL, Jolesz FA, Brookeman JR. Optimized single-slab three-dimensional spin-echo MR imaging of the brain. Radiology. 2000;216:891–899.
11. Busse RF, Hariharan H, Vu A, Brittain JH. Fast spin echo sequence with very long echo trains: design of variable refocusing flip angle schedules and generation of clinical T2contrast. Magn Reson Med. 2006;55:1030–1037.
12. Otazo R, Candes E, Sodickson DK. Low-rank plus sparse matrix decomposition for accelerated dynamic MRI with separation of background and dynamic components. Magn Reson Med. 2015; 73: 1125 1136.
13. Trémoulhéac, B, Nikolaos D, David A, and Simon RA. Dynamic MR Image Reconstruction–Separation From Undersampled k-t Space via Low-Rank Plus Sparse Prior." IEEE Trans Med Imaging 2014; 33: 1689-1701.
14. Petrov, AY, Herbst, M, and Stenger, VA. Improving temporal resolution in fMRI using a 3D spiral acquisition and low rank plus sparse (L+ S) reconstruction. NeuroImage, 2017; 157: 660-674.
Figures
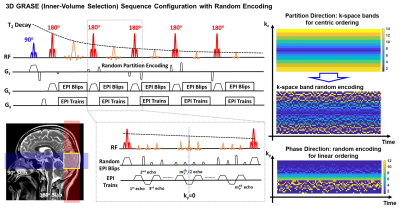
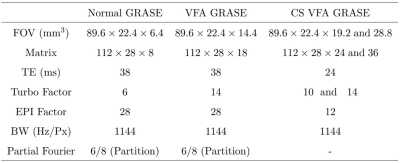
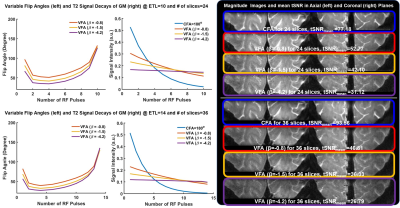
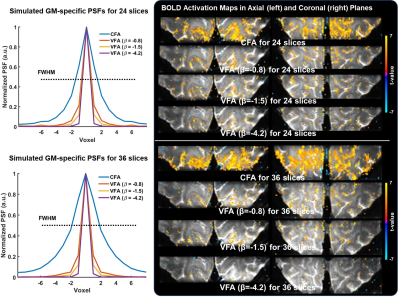
Fig. 3. Gray matter specific simulated PSFs under different regimes of constant flip angle (CFA) and VFA, and the corresponding BOLD activation maps in the axial and coronal planes for both 24 and 36 slices. Consistent with the finding from the PSF simulations, the CFA activation maps introduces BOLD smearing effect across neighboring regions, while the VFAs improve spatial specificity at the cost of sensitivity.
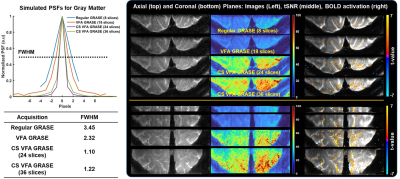
Fig. 4. Comparison of GM-specific PSFs, magnitude images, tSNR maps, and BOLD activation maps in the axial and coronal planes for normal, VFA, and CS VFA GRASE (24 and 36 slices). Note that proposed CS VFA GRASE outperforms normal and VFA GRASE in terms of image quality, tSNR, and BOLD sensitivity even with a sharper PSF.