1050
Longitudinal FreeSurfer with non-linear subject-specific template improves sensitivity to cortical thinning1Athinoula A. Martinos Center for Biomedical Imaging, Charlestown, MA, United States, 2Department of Radiology, Harvard Medical School, Boston, MA, United States, 3German Center for Neurodegenerative Diseases, Bonn, Germany, 4Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA, United States
Synopsis
Longitudinal FreeSurfer creates a within-subject template by rigidly registering and median-filtering longitudinal timepoints (TP). Information common to all TPs is extracted from the template for unbiased TP initialization, resulting in substantial improvements over cross-sectional processing. However, this approach is not optimal in the presence of severe atrophy or other large-scale anatomical change, which causes voxels to be filtered across tissue classes. We address this problem by introducing an enhanced longitudinal stream that deforms each TP using non-linear registration to construct the template. We demonstrate considerable increases in sensitivity to cortical thinning, without affecting test-retest reliability.
Authorship
*Sharing senior authorship.Introduction
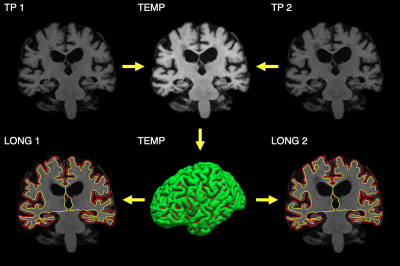
Longitudinal design has become increasingly popular in studies into aging and disease-related brain atrophy due to increased sensitivity to temporal change. This trend led to the development of processing pipelines tailored to longitudinal datasets such as longitudinal FreeSurfer (FS)1. In FS, a subject-specific template is created by rigidly registering and median-filtering timepoints (TP)2, as explained in Figure 1. This allows the extraction of information common to all TPs to initialize the processing of each TP without treating any one of them (e.g. the baseline) differently than the others, to avoid introducing processing bias.1While this approach resulted in substantial improvements over cross-sectional processing1, it is not optimal in the presence of large scale changes in anatomy across time. Figure 2 illustrates that creating the base from two rigidly aligned TPs can be equivalent to filtering voxels across tissue classes. Our contribution overcomes this problem by introducing a non-linear coordinate system into the template creation. We present and evaluate an enhanced longitudinal stream which deforms each TP using non-linear registration to construct the template.
Methods
Subject-specific templateTemplates were generated using ANTs'3 multivariate template creation, modified to perform non-linear SyN3 registration only. A rigid template was obtained as the median of normalized skull-stripped images from standard FS after robust registration2 into an unbiased mid-space, as in longitudinal FS1. Iterative deformable registration of each TP to the template was initialized with the rigid transforms, and the template was updated after each iteration (n=4). We computed the final template as the voxel-wise median of the warped TPs.
FreeSurfer integration and processing
The template creation was integrated with FS/recon-all for longitudinal processing. White/pial surfaces were generated for the non-linear template and copied to each TP for initialization. Longitudinal processing was performed in the space of the initial rigid template since warping TPs into non-linear-template space would remove effects. Template surfaces were not deformed with the warp fields for TP initialization.
Sensitivity to cortical atrophy
Sensitivity to cortical thinning was evaluated in n=50 stable Alzheimer’s disease (AD) patients and 50 controls (CN) at 0 and 24 months. We selected most likely healthy/diseased CN/AD subjects based on a score attributing one point for zero/each APEO e4 allele copy, below/above median ptau and tau and above/below median abeta. Median values were computed for subjects with mild cognitive impairment. T1-weighted 3D scans at 1.5T were obtained from the Alzheimer’s Disease Neuroimaging Initiative database (ADNI14) and processed using cross-sectional/longitudinal FS and the proposed non-linear stream. Group-wise annual atrophy rates were fitted5 for labels of the Desikan-Killiany atlas6, controlling for sex, age and with random intercepts for each subject.
Test-retest reliability
We assessed the test-retest reliability of cortical thickness measures in T1-weighted back-to-back scans of n=50 subjects randomly chosen from the MIRIAD database7. Repeat acquisitions were not contingent on subject compliance. Imaging was performed at 1.5T. For each subject, differences in thickness ti (i=1,2) were summarized as absolute symmetrized percent change:
$$\Delta t = 2 \frac{ \mid t_2 - t_1 \mid}{t_2 + t_1} \times 100$$
Results
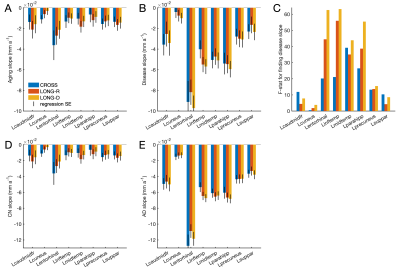
Sensitivity to cortical atrophyFigure 3A-B shows aging and disease slopes obtained with each pipeline for structures in the left cortex. Atrophy rates were of the order of a few 10-2 mm/year and varied across structures. The non-linear stream tended to produce lower aging and higher disease slopes than longitudinal FS. This boosted statistical power for structures associated with AD, e.g. entorhinal cortex and the parahippocampal gyrus, as reflected in the F-statistic in Figure 3C.
Test-retest reliability
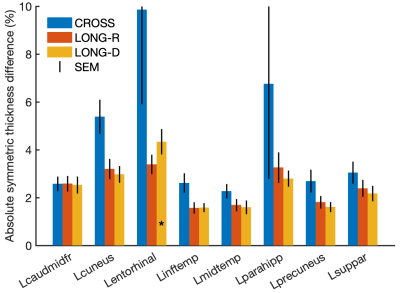
Test-retest differences for each stream are compared in Figure 4. As expected, longitudinal FS and the new stream had substantially lower variability than cross-sectional FS. Thickness differences for the new stream were similar to those found with longitudinal FS (typically 2-4%).
Longitudinal surface placement
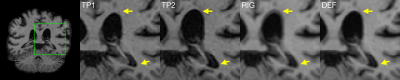
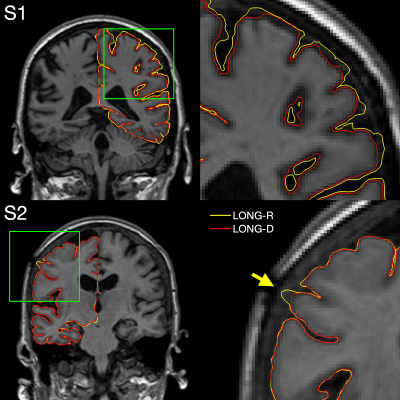
In a few ADNI1 subjects, processing the non-linear template resulted in improved surface placement as compared to the rigid template, e.g. if dura was mistakenly included in the pial surface. This can be seen in Figure 5, which compares pial surfaces for both streams.
Discussion
We demonstrated substantial increases in sensitivity to cortical thinning using a non-linear subject-specific template with longitudinal FS, without affecting test-retest reliability. These improvements may be useful for the early detection of disease or in studies with limited numbers of subjects.With runtimes of t~2h per SyN registration on a 3.3-GHz Intel Xeon CPU, creating the template with n=4 iterations took about 16h for two TPs. This computational burden requires parallelization for the analysis of large datasets. Further work may exploit the efficiency of deep learning, e.g. by estimating deformation fields with VoxelMorph8.
Initial experiments exploring template surface warping for TP initialization resulted in sensitivity losses, presumably because surface optimization operates along face normals and is highly depended on the starting point. Further work is needed to fully leverage the information in the deformation fields.
Conclusion
We introduced a non-linear coordinate system into the within-subject template of longitudinal FS and demonstrated improved sensitivity to cortical atrophy, without affecting test-retest reliability. This contribution may be useful to users who seek to detect early changes in disease or recruit fewer subjects.Acknowledgements
Support for this research was provided in part by grant NIH U01 AG052564, by the BRAIN Initiative Cell Census Network grant U01MH117023, the National Institute for Biomedical Imaging and Bioengineering (P41EB015896, 1R01EB023281, R01EB006758, R21EB018907, R01EB019956), the National Institute on Aging (1R56AG064027, 5R01AG008122, R01AG016495), the National Institute of Mental Health the National Institute of Diabetes and Digestive and Kidney Diseases (1-R21-DK-108277-01), the National Institute for Neurological Disorders and Stroke (R01NS0525851, R21NS072652, R01NS070963, R01NS083534, 5U01NS086625,5U24NS10059103, R01NS105820), and was made possible by the resources provided by Shared Instrumentation Grants 1S10RR023401, 1S10RR019307, and 1S10RR023043. Additional support was provided by the NIH Blueprint for Neuroscience Research (5U01-MH093765), part of the multi-institutional Human Connectome Project. In addition, BF has a financial interest in CorticoMetrics, a company whose medical pursuits focus on brain imaging and measurement technologies. BF's interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.;Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.;Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
References
1. Reuter M, Schmansky NJ, Rosas HD, Fischl B. Within-subject template estimation for unbiased longitudinal image analysis. Neuroimage. 2012;61(4):1402-1418.
2. Reuter M, Rosas HD, Fischl B. Highly accurate inverse consistent registration: A robust approach. Neuroimage. 2010;53(4):1181-1196.
3. Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008;12(1):26-41.
4. Weiner M. Alzheimer’s Disease Neuroimaging Initiative. https://adni.loni.usc.edu. Published 2003. Accessed November 5, 2019.
5. Bernal-Rusiel
JL, Greve DN, Reuter M, Fischl B, Sabuncu MR. Statistical Analysis of
Longitudinal Neuroimage Data with Linear Mixed Effects Models. Neuroimage.
2013;1:249.
6. Desikan RS, Ségonne F, Fischl B, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31(3):968-980.
7. Malone IB, Cash D, Ridgway GR, et al. MIRIAD—Public release of a multiple time point Alzheimer’s MR imaging dataset. Neuroimage. 2013;70:33-36.
8. Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV. VoxelMorph: A Learning Framework for Deformable Medical Image Registration. IEEE Trans Med Imaging. 2019;38(8):1788-1800.
Figures