1048
MRI Raw Data Compression for long-term storage in large-scale population imaging1German Center for Neurodegenerative Diseases (DZNE), Bonn, Germany
Synopsis
In the overwhelming majority of MRI studies, only the reconstructed images are stored and the raw data that was used during the reconstruction process is lost. However, routine raw data storage would potentially allow imaging studies that are conducted now to benefit from future improved image reconstruction techniques. Unfortunately, the raw data storage requirements are often prohibitive, especially in large-scale population studies. We developed a flexible software tool that achieves high lossy compression of MRI raw data and is able to decompress the data back to the vendor-specific format to allow for retrospective reconstruction using the vendor's reconstruction pipeline.
Introduction
In the overwhelming majority of MRI studies, only the reconstructed images are stored and the raw data that was used during the reconstruction process is lost. However, routine raw data storage would potentially allow imaging studies that are conducted now to benefit from future developments: Reconstruction techniques evolve quickly and new techniques often provide significantly better image quality or even new information. Furthermore, it is often much easier to mitigate or correct for image artifacts in the raw data than in the reconstructed images, which is only possible if the raw data is still available after the image artifact has been detected.The Human Connectom Project [1] is a large project with over 1000 participants that already benefited from the routine storage of the MRI raw data: The dMRI and fMRI image reconstruction was improved a few years after the start of the project and then retrospectively applied to the data that was acquired up to that point. For even larger population studies, such as the UK Biobank imaging study [2] or the Rhineland Study [3], routine storage of the raw data would be very challenging and expensive due to the huge amount of data. Thus, compression techniques that significantly reduce the amount of data are essential. Since lossless compression of the noisy raw data is very limited, efficient lossy compression techniques are required that provide high compression factors while still retaining most of the important information. The aim of this project is to develop a flexible software tool for lossy compression of MRI raw data and that decompresses the data back to the vendor-specific format to allow for retrospective reconstruction using the vendor's reconstruction pipeline.
Methods
A 3D T2-weighted image was acquired from a healthy subject on a 3T system (Siemens Prisma) using a 64 channel head coil array (with 58 channels active) and the raw data (7 GB in vendor-specific format) was stored along with the DICOM images. The raw data file was then compressed with various lossy compression techniques using a custom python-based software tool.The compression tool first separates the protocol header as well as meta data information from the imaging data itself. In a first step, lossless compression (gzip) is applied to the header and meta data information which is then added to a hdf5 file [4]. Then, before adding the data, different lossy compression techniques are applied (as well as combinations thereof), namely: a) downsampling in order to remove the standard 2x ADC oversampling, b) lossy floating-point compression using the zfp library [5], c) svd-based coil compression, either "single coil compression" (scc) or "geometric coil compression" (gcc) [6]. All additional meta information necessary for decompression (e.g. coil compression matrices) are also stored in the hdf5 file.The compression factor was determined by dividing the byte size of the original raw data file by the size of the hdf5 file. Using the same tool, the file was then decompressed back to the vendor format (resulting in the original byte size) and was brought back to the scanner for retrospective reconstruction. Several image quality metrics were calculated from the original image and the images that were retrospectively reconstructed from the compressed data.Results
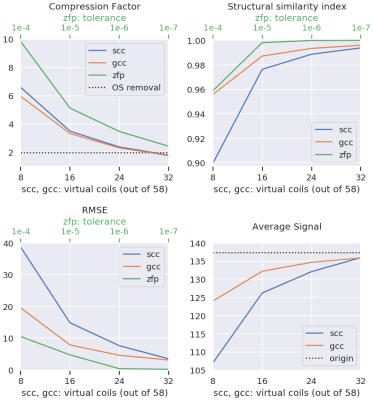
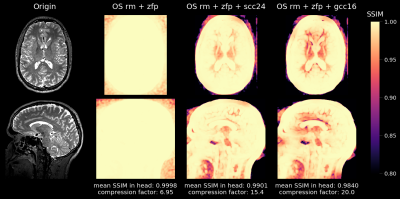
Figure 1 shows achievable compression factors as well as quality metrics for the different compression techniques. As expected, all quality metrics degrade at higher compression factors. The structural similarity metric (SSIM) results of three different compression technique combinations (overall compression factor 6.95-20.0) are presented in Figure 2.Discussion
We present a software tool for (Siemens) MRI raw data compression and decompression that uses various lossy compression techniques, namely oversampling removal, floating-point compression, and coil compression. The preliminary results based on the analysis of a 3D T2-weighted scan indicate that compression factors between 7 and 20 are feasible. However, for long term storage, it may be advisable to err on the side of caution since it is difficult to foresee how a possible future reconstruction techniques will react. But in the end, the most appropriate compression factor will depend on a tradeoff between information retention and storage costs.Acknowledgements
Financial support of the Helmholtz Association (Helmholtz Incubator pilot project “Imaging at the Limits”) is gratefully acknowledged.References
[1] Marcus DS, , et al. Human Connectome Project informatics: quality control, database services, and data visualization. Neuroimage. 2013;80:202-219.
[2] https://imaging.ukbiobank.ac.uk
[3] https://www.rheinland-studie.de
[4] The HDF Group. Hierarchical Data Format, version 5, 1997-2019. http://www.hdfgroup.org/HDF5/.
[5] Peter Lindstrom, Fixed-Rate Compressed Floating-Point Arrays. IEEE Trans Vis Comput Graph. 2014;20:2674-2683.
[6] Zhang T, et al. Coil compression for accelerated imaging with Cartesian sampling. Magn Reson Med 2013;69:571-582.
Figures