1046
A Reconstruction Compatible, Fast and Memory Efficient Visualization Framework for Large-scale Volumetric Dynamic MRI1Radiology, Stanford University, Stanford, CA, United States, 2Electrical Engineering, Stanford University, Stanford, CA, United States, 3Electrical and Computer Sciences, UC Berkeley, Berkeley, CA, United States, 4Electrical and Computer Engineering, The University of Texas at Austin, Austin, TX, United States
Synopsis
We addressed the speed and memory shortcomings of conventional visualization consoles when processing high-dimensional MRI datasets by proposing a novel approach that leverages compressed representations of such datasets. We considered low rank reconstructions and operated on them directly for visualization, unlike traditional viewers which load entire uncompressed image datasets. We built a web viewer that utilizes this approach to demonstrate real time reformatting and slicing. We were able to achieve more than 15x reduction in both memory usage and loading times.
Introduction
High-dimensional MRI has become an important component in many applications, such as dynamic contrast enhanced imaging1, 4D flow2, and fast spin echo knee imaging3. Among its many advantages, 3D dynamic MRI in particular offers high quality multiplanar and temporal reformatting, which can greatly enhance clinical interpretation. Recent advances in compressed sensing and low rank methods have further pushed the achievable spatio-temporal resolution limits. These reconstruction techniques are now able to reconstruct large-scale volumetric dynamic datasets by exploiting the underlying low dimensional image representation.On the other hand, visualizing and storing these large sets of images pose immense challenges to hospital computing backends. Datasets stored in DICOM formats often have sizes on the order of 10-100 GBs, which result in expensive storage costs and slow data transfer. Moreover, visualization consoles require massive amounts of memory just to load the images. This can cause latency in visualization as long as 10 seconds per reformat, which impedes radiologists’ workflow. While off-the-shelf lossless compression methods can reduce data sizes, they further increase loading times.
In this work, we propose a fast and memory-efficient framework for visualizing volumetric dynamic MRI by exploiting abstract representations used in reconstructions. In particular, instead of storing reconstructed images in DICOM formats, we leverage low rank representations already used in reconstruction and store them directly in a lossless format. We show that reformatting and slicing such data can be done in real time, thereby eliminating the need to operate on the full image dataset. We demonstrate the effectiveness of the proposed visualization framework on two low rank based reconstruction methods, T2 shuffling4 and multi-scale low rank5, and show that memory usage and latency can be reduced by >10x.
Methods
While our framework can be applied to general compressed sensing reconstructions, we consider low rank reconstruction methods for brevity. In particular, for $$$T$$$ frames, we consider low rank reconstruction methods that produce spatial basis vectors ($$$L \in \mathbb{C}^{N \times K}$$$) and temporal basis vectors ($$$R \in \mathbb{C}^{T \times K}$$$), such that they are related to the underlying dynamic image series $$$X = [x_1, \ldots, x_T]$$$ by,$$X = L R^H$$
We note that by defining the support of the basis vectors appropriately, the above relationship includes general structured low rank models, such as local low rank6 and multi-scale low rank5.
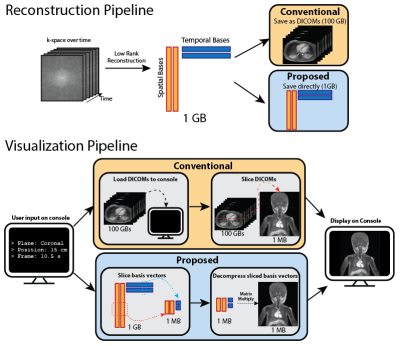
To constrain the reconstruction problem, low rank methods prescribes the matrix rank $$$K$$$ to be much less than the number of frames $$$T$$$. This property allows for a dramatic reduction in memory footprint. For example, for an 80-frame dynamic image sequence, a rank of 4 attains a 20x compression factor. Hence, instead of storing the full image set as DICOMs, we propose saving and operating on the compressed representation directly to maintain memory efficiency.
Moreover, the low rank compressed representation allows us to slice and reformat a particular slice $$$n$$$ and frame $$$t$$$ efficiently without ever operating on the full image set. In particular, we can slice the spatial basis vector $$$L[n]$$$ and the temporal basis vector $$$R[t]$$$ independently. The desired slice is then obtained through a matrix multiplication of the sliced basis vectors:
$$L[n] R[t] ^H$$
The full pipeline is described in Figure 1.
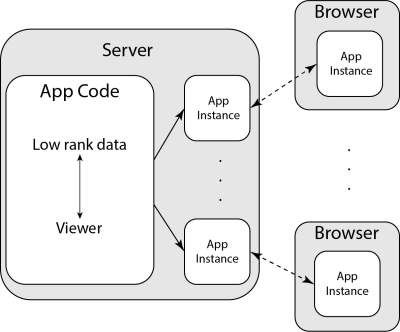
For visualization, we built a viewer capable of accessing and interacting with low rank reconstructed datasets using the above procedure. The viewer uses a Python based interactive visualization library, Bokeh7. The application code for the viewer is hosted on a server, which creates application instances upon request, shown in Figure 2. This architecture allows for simple deployment for a network of users who can simultaneously interact with the application.
Results
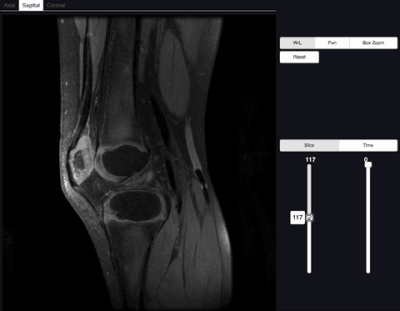
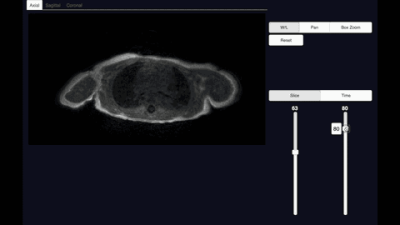
We demonstrated our visualization software on a knee dataset reconstructed with T2 Shuffling (Figure 3). The dataset when uncompressed has a matrix size of 260x240x288 with 80 time frames, and requires ~10.8GBs to store. In contrast, the compressed representation only used 660MB. Loading the uncompressed data took ~10s, whereas our proposed method took ~0.5s on a laptop.We also applied the proposed method on a chest DCE dataset reconstructed with the multi-scale low rank reconstruction method (Figure 5). The dataset if uncompressed has a matrix size of 323x186x332 with 500 time frames and takes ~74GBs, which cannot be loaded in common viewer consoles. In contrast, the compressed representation only took ~1.7GBs and can be reformatted in real-time.
Conclusion
The main advantage of our method over existing visualization methods is that our method leverages the compressed representation used in reconstruction and does not require the fully memory footprint to obtain slices of the image. Our method allows for better ease of access to large scale MRI datasets by enabling support for web applications. The more responsive visualization has the potential to greatly benefit the clinical workflow.Acknowledgements
No acknowledgement found.References
1. Hamm, B., Thoeni, R. F., Gould, R. G., Bernardino, M. E., Lüning, M., Saini, S., Mahfouz, A. E., Taupitz, M. and Wolf, K. J. (1994). Focal liver lesions: characterization with nonenhanced and dynamic contrast material-enhanced MR imaging. Radiology, 190(2), 417–423. doi: 10.1148/radiology.190.2.8284392
2. Markl, M. , Frydrychowicz, A. , Kozerke, S. , Hope, M. and Wieben, O. (2012), 4D flow MRI. J. Magn. Reson. Imaging, 36: 1015-1036. doi:10.1002/jmri.23632
3. Bredella, M. A., Tirman, P. F., Peterfy, C. G., Zarlingo, M. F., Feller, J. W., Bost, F. P., Belzer, J.P., Wischer, T.K. and Genant, H. K. (1999). Accuracy of T2-weighted fast spin-echo MR imaging with fat saturation in detecting cartilage defects in the knee: comparison with arthroscopy in 130 patients. American Journal of Roentgenology, 172(4), 1073–1080. doi: 10.2214/ajr.172.4.10587150
4. Tamir, J. I., Uecker, M. , Chen, W. , Lai, P. , Alley, M. T., Vasanawala, S. S. and Lustig, M. (2017), T2 shuffling: Sharp, multicontrast, volumetric fast spin‐echo imaging. Magn. Reson. Med., 77: 180-195. doi:10.1002/mrm.26102
5. Ong, F. and Lustig, M. (2016). Beyond Low Rank + Sparse: Multi-scale Low Rank Matrix Decomposition. IEEE journal of selected topics in signal processing, 10(4), 672–687. doi:10.1109/JSTSP.2016.2545518
6. Trazasko, J. D. (2013). Exploiting local low-rank structure in higher-dimensional MRI applications. In Wavelets and Sparsity XV (Vol. 8858). [885821] https://doi.org/10.1117/12.2027059
7. Bokeh Development Team (2018). Bokeh: Python library for interactive visualization URL http://www.bokeh.pydata.org.
Figures