1039
Virtual Scanner: MRI Experiments in a Browser1Department of Biomedical Engineering, Columbia University, New York, NY, United States, 2Columbia Magnetic Resonance Research Center, Columbia University, New York, NY, United States
Synopsis
Open-source standards for MR pulse sequences and data have been recently developed, but there is no unified platform for combining them with implemented simulation, reconstruction, and analysis tools to the best of our knowledge. We designed Virtual Scanner in order to provide a platform that allows rapid prototyping of new MR software and hardware. It also serves as a training tool for MR technicians and physicists. Two modes are provided: Standard Mode mimics MR scanner interfaces to assist training, while Advanced Mode allows customized simulation of each step in the signal chain.
Introduction
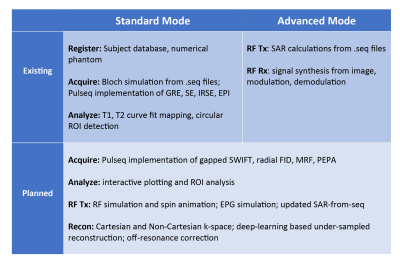
Open-source standards such as Pulseq1 and ISMRMRD2 have been developed to facilitate vendor-neutral research. To make the best use of them, an integrated platform for simulation, reconstruction, and analysis tools is necessary. We developed Virtual Scanner to be this platform so that researchers can share their methods more efficiently and validate them across MR system manufacturers. The same tools, on the other hand, also serve education purposes when connected to the console-like user interface.Virtual Scanner has two modes (Figure 1). The Standard Mode mimics the workflow on a clinical MR console to help train MR technicians, while the Advanced Mode contains elements of the signal chain for conducting MR research and training MR engineers.
Methods
We programmed Virtual Scanner in Python 3.6 and built the browser user interface with FLASK.3 A payload dictionary is sent between the client and the server to exchange information such as user-input parameters and files.In Standard Mode, a subject database stores user and phantom information, a Bloch simulator generates the signal from a numerical phantom and a Pulseq MR sequence file, and a multiparameter curve fitting module maps out T1 and T2 from series of acquisitions. The sequence file can be generated either with the original MATLAB implementation or the PyPulseq package.4 The simulator takes inputs of .seq files and applies Bloch equation time-stepping to obtain signal summed from all isochromats.5 Each isochromat is defined with a location, proton density, T1, and T2. Multiparameter curve fitting generates T1 and T2 maps from series of images acquired with different parameters.
In Advanced Mode, the Tx page calculates the Specific Absorption Rate (SAR) directly from Pulseq sequences using the Q-matrix method,6 and the Rx page synthesizes time-domain MR signal from images and can perform modulation, demodulation, and reconstruction.
These two modes are separated at login with two distinct groups of users in mind: the MR technician and the MR engineer. A multi-tab design for each mode with input fields on the bottom and display sections on top resembles typical MR console layout (Figure 1). Display of multiple simulated images in the axial, sagittal, and coronal planes is possible. Data loading is also enabled for Tx, where the user may check SAR limits using their own .seq file.
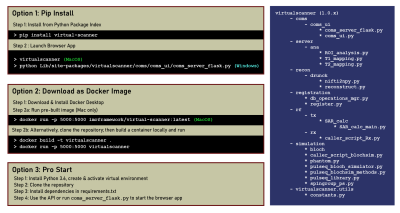
Three options are provided for deployment: (1) Pip installation from the Python Package Index; (2) Using a Docker image; (3) Cloning the repository. Users may choose depending on the amount of Python code with which they wish to interact (Figure 2). Detailed instructions can be found on the Github repository.7
Results
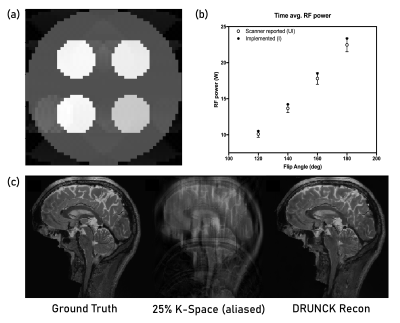
The Standard Mode allows walk-through of the MRI scan: registering the patient, choosing a sequence, setting parameters, performing a scan, and generating T1 and T2 maps from images. The prototypical Advanced Mode demonstrates the signal processing steps from time-domain signal to image and generates SAR plots that are key in assessing the practical value of custom sequences. Apart from GUI-based interaction, we also use the modules for simulating and validating new acquisition methods and incorporate scripts back into Virtual Scanner.Figure 3 shows examples of features in development. A parametrized Echo Planar Imaging (EPI) sequence is developed with options of Spin Echo versus Gradient Echo, the number of shots, and blocked or interleaved trajectories. The updated SAR4seq feature helps test the feasibility of custom Pulseq sequences before playing them on a scanner. The Deep Learning Reconstruction of Under-sampled Cartesian K-space (DRUNCK) method is a more advanced addition to the data loading and standard reconstruction features.
We are actively developing new backend features shown in Figure 4 to facilitate both kinds of usage. These features add to the library of pulse sequences, reconstruction methods, and analysis options. MR researchers can contribute and provide feedback through Github community features such as pull requests and issue posts.
Discussion
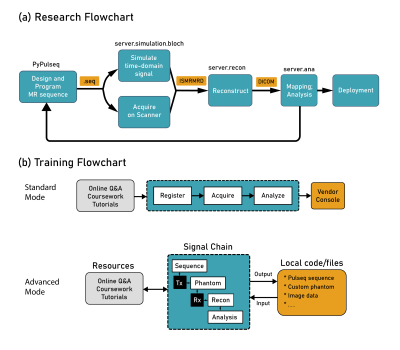
Virtual Scanner is extensively documented for the MR community to use in their research.8 A research pipeline can be set up by installing the Pulseq interpreter on the scanner, generating a new Pulseq sequence, and then testing it with Virtual Scanner simulation or scanner data acquisition (Figure 5a). Both simulated and real signals can be fed through reconstruction, signal analysis, and quantitative mapping, completing a feedback loop for rapid development of novel MR methods. More functions, such as Pulseq sequence plotting, SAR calculations, and user-made code, are needed to facilitate this process and are being developed.Virtual Scanner also supports MR education (Figure 5b). The console-like interface can help familiarize MR technicians with running protocols and reduces the number of training hours on a real scanner. MR physicists can utilize the more advanced features by interacting through the user interface, scripting, and custom files such as sequences, phantom parameters, and images.
Conclusion
We designed Virtual Scanner not only to be a free research tool, but also to lower the barrier to accessing and maintaining MR technology deployment in underserved regions, by emphasizing open-source standards and system-wide coverage. Future work will focus on GUI integration and new simulation, reconstruction, and visualization features to complete the signal chain.Acknowledgements
1. Zuckerman Institute Technical Development Grant for MR, Zuckerman Mind Brain Behavior Institute,Grant Number: CU-ZI-MR-T-0002; PI: Geethanath
2. Zuckerman Institute Seed Grant for MR studies, Zuckerman Mind Brain Behavior Institute,Grant Number:CU-ZI-MR-S-0007; PI: Geethanath
References
1. Layton, K. J., Kroboth, S., Jia, F., Littin, S., Yu, H., Leupold, J., ... & Zaitsev, M. (2017). Pulseq: a rapid and hardware‐independent pulse sequence prototyping framework. Magnetic resonance in medicine, 77(4), 1544-1552.
2. Inati, S. J., Naegele, J. D., Zwart, N. R., Roopchansingh, V., Lizak, M. J., Hansen, D. C., ... & Xue, H. (2017). ISMRM Raw data format: A proposed standard for MRI raw datasets. Magnetic resonance in medicine, 77(1), 411-421.
3. Pallets. (n.d.). Pallets/flask. GitHub. Retrieved from https://github.com/pallets/flask
4. Ravi et al., (2019). PyPulseq: A Python Package for MRI Pulse Sequence Design. Journal of Open Source Software, 4(42), 1725, https://doi.org/10.21105/joss.01725
5. Kose, R., & Kose, K. (2017). BlochSolver: A gpu-optimized fast 3D mri simulator forexperimentally compatible pulse sequences. Journal of Magnetic Resonance, 281, 51–65. doi:10.1016/j.jmr.2017.05.007
6. Graesslin, I., Homann, H., Biederer, S., Börnert, P., Nehrke, K., Vernickel, P., Mens, G., et al. (2012). A specific absorption rate prediction concept for parallel transmission MR. Magnetic Resonance in Medicine, 68(5), 1664–1674. doi:10.1002/mrm.24138
7. imr-framework/virtual-scanner. (2019). Retrieved 5 November 2019, from https://github.com/imr-framework/Virtual-Scanner
8. Welcome to Virtual Scanner's documentation! (2019). Retrieved 6 November 2019, from https://imr-framework.github.io/virtual-scanner/.
Figures