1036
End-to-End Full Automated Pipeline using a Convolutional Neural Network for Lung Segmentation in Phase-Resolved Functional Lung (PREFUL) MRI1Institute of Diagnostic and Interventional Radiology, Hanover Medical School, Hannover, Germany, 2Biomedical Research in Endstage and Obstructive Lung Disease Hannover (BREATH), German Center for Lung Research (DZL), Hannover, Germany, 3Biomedical Research in Endstage and 3 Obstructive Lung Disease Hannover (BREATH), German Center for Lung Research (DZL), Hannover, Germany
Synopsis
Translation and establishment of complex pulmonary magnetic resonance (MR) imaging techniques in the clinics requires a reliable, fully automated and fast calculation. In this work we present a semantic convolutional neural network (CNN) model for lung parenchyma and vessel segmentation in combination with parallelized computation on a high-performance computer to design an end-to-end pipeline for phase-resolved functional lung (PREFUL) MRI. The CNN was trained (n=1118) and validated (n=1064) with manually segmented images by a trained radiologist. Automatic segmentation of lung parenchyma was achieved for all tested images.
Synopsis
Translation and establishment of complex pulmonary magnetic resonance (MR) imaging techniques in the clinics requires a reliable, fully automated and fast calculation. In this work we present a semantic convolutional neural network (CNN) model for lung parenchyma and vessel segmentation in combination with parallelized computation on a high-performance computer to design an end-to-end pipeline for phase-resolved functional lung (PREFUL) MRI. The CNN was trained (n=1118) and validated (n=1064) with manually segmented images by a trained radiologist. Automatic segmentation of lung parenchyma was achieved for all tested images.Introduction
Convolutional Neural Networks (CNN) have drastically broadened the potential of computational models to represent data of multiple levels of abstraction [1]. In the field of radiology CNNs have been used for several applications such as classification [2], detection [3], segmentation [4] and image reconstruction [5]. CNNs have shown to enable automated diagnosis at high data throughput and improved accuracy [6] in a clinical setting.Phase-resolved functional lung (PREFUL) MR imaging allows pulmonary ventilation and perfusion assessment without the use of any radiation or contrast agent at high patient compliance [7]. However, a time-consuming workflow in the post-processing of PREFUL due to manual lung segmentation and costly computationally steps like the image registration, impede its extensive application in clinical routine. The purpose of this work was to automatize the lung parenchyma and vessel segmentation using a semantic CNN, the parallelization of the registration process and implementing the PREFUL algorithm on a high-performance computer.
Methods
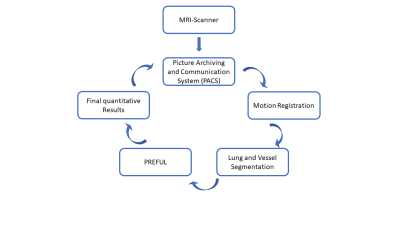
All computations were performed on a high-performance computer (X11DPG-QTMainboard, LGA 3647, 2 x Xeon Platinum 8176 2.1GHz 112-Core, 256GB Ram, 2 x NVIDIA Quadro RTX 6000 24 GB). 1118 coronal MR images from COPD patients of a single centre study [8] were manually segmented defining the ground truthas input parameters. The MR images were acquired a 1.5T MR-scanner (Magnetom Avanto, Siemens Healthcare, Erlangen, Germany) as described in detail, previously [8]. In the training phase, a pre-trained CNN for classification and detection (VGG16) [9] was selected and extended to our needs to automatically detect the lung parenchyma. To validate the accuracy of the segmented ROIs 1064 additional manually segmented MR images were used. The scaling geometric factor [10] and the Dice [11] similarity coefficient for image segmentation were calculated to assess the performance of CNN network.The fully automated pipeline consisted of three main steps, see Figure 2. The first part comprised of the MR images import from the Picture Archiving and Communication System (PACS) to the server and the registration of the images. The image registration was performed with the freely available Advanced Normalization Tools (ANTs) [12] using the group-oriented registration approach [13]. The images were retrieved from PACS using defined keywords to find the appropriate data sets. Then, the slice registrations were divided among the different CPU kernels, which resulted in a total calculation duration of one hour for 56 slices with 250 images per slice. Once the registration is done, the CNN provided 56 ROIs in 11 s. For comparison, a standard office PC, (for example an Intel Iris Pro 1536 MB, 16GB, 1600 MHz DDR3 2,2 GHz Intel Core i7), would need approximately 56 hours for the registration of 56 slices and an experienced radiologist requires 60 min for manual segmentation of 56 slices.
Results
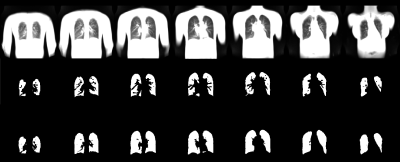
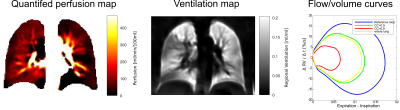
Exemplary results of the manual and automatic segmentation are shown in Figure 1. A segmentation performance with a scaling geometric factor of 3.7 corresponding to a Dice coefficient of 78.3% of accuracy was achieved in the comparison of manual and automated segmentation. Figure 2 shows the work-flow of the fully automated program. The results of the lung parenchyma and large vessel segmentation can be seen in figure 3. Figure 4 shows the final PREFUL extracted parameters of a 19 years old cystic fibrosis (CF) patient.Discussion
The overlap obtained between manual segmentation and automated segmentation was moderate and affected as follows: For implementation in the PREFUL pipeline an automated segmentation of main pulmonary vessels was desired. Therefore, the CNN network was trained to segment lung parenchyma without large vessels in comparison to manual segmentation, where also smaller vessels were excluded (see Figure 1). A further obstacle for successful automated segmentation might be given by morphological changes of the lung parenchyma in patients, which might not be recognized by our CNN network. Therefore, the performance of a trained CNN network may be increased in short amount of time. Previously, pipelines for free breathing 1H proton lung MRI measurement including automated segmentation have been developed [14]. With the usage of 56 kernels on a high-performance computer, the registration time was minimized by a factor of 56. The registration step is further limited by the fact, that ANTs does not enable image registration on GPU. Therefore, in the future, image registration may be further accelerated by applying artificial intelligence.Conclusions
This work shows a successful combination of CNN for pulmonary segmentation and the parallelization of extensive computations to establish an end-to-end pipeline for image registration and PREFUL analysis, which can be used to obtain fully automated pulmonary ventilation and perfusion information in a reasonable time-frame.Acknowledgements
This work was funded by the German Center for Lung Research (DZL).References
1. Yamashita, R., Nishio, M., Do, R.K.G. et al. Insights Imaging (2018) 9: 611. https://doi.org/10.1007/s13244-018-0639-9
2. Yasaka K, Akai H, Abe O, Kiryu S (2018) Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: a preliminary study. Radiology 286:887–896
3. Lakhani P, Sundaram B (2017) Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology 284:574–582
4. Christ PF, Elshaer MEA, Ettlinger F et al (2016) Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields. In: Ourselin S, Joskowicz L, Sabuncu M, Unal G, Wells W (eds) Proceedings of Medical image computing and computer-assisted intervention – MICCAI 2016.
5. Kim KH, Choi SH, Park SH (2018) Improving arterial spin labeling by using deep learning. Radiology 287:658–666.
6. Ardila, D:, Kiraly, A.P., Bharadwaj, S. et al. End-to-End lung cancer screening with three dimensional deep learning on low-dose chest computed tomography. Nat Med 25, 954-961 (2019) doi: 10.1038/s41591-019-0447-x
7. Voskrebenzev A, Klimes F, Gutberlet M, et al. Feasibility of quantitative regional ventilation and perfusion mapping phase-resolved functional lung (PREFUL) MRI in healthy volunteers and COPD, CTEPH, and CF patients. Magn Reson Med. 2018; 79(4):2306-2314. doi: 10.1002/mrm.26893.
8. Am J Respir Crit Care Med. 2019 May 1;199(9):1086-1096. doi: 10.1164/rccm.201805-0995OC.
9. Simonyan K, Zisserman A. Very Deep Convolutional Neural Networks for Large-Scale Image Recognition. International Conference on Learning Representations. 2015.
10. Stegmann, M. B., & Delgado, D. (2002). A brief introduction to Statistical Shape Analysis. Informatics and Mathematical Modelling, (March), 15 pp.
11. Zou KH, Warfield SK, Bharatha A, et al. Statistical validation of image segmentation quality based on a spatial overlap index. Acad Radiol. 2004;11(2):178–189. doi:10.1016/s1076-6332(03)00671-8
12. Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54(3):2033-2044. doi:10.1016/j.neuroimage.2010.09.025
13. Voskrebenzev A, Gutberlet M, Kaireit TF, Wacker F, Vogel-Claussen J. Low-pass imaging of dynamic acquisitions (LIDA) with a group-oriented registration (GOREG) for proton MR imaging of lung ventilation. Magn Reson Med2016. doi: 10.1002/mrm.26526.
14. Guo F, Capaldi DPI, McCormack DG, Fenster A, Parraga G. A framework for Fourier-decomposition free-breathing pulmonary 1 H MRI ventilation measurements. Magn Reson Med. 2018;(August):1-12. doi:10.1002/mrm.27527
Figures