1035
SARDU-Net: a new method for model-free, data-driven experiment design in quantitative MRI1Queen Square MS Centre, Queen Square Institute of Neurology, Faculty of Brain Sciences, University College London, London, United Kingdom, 2Centre for Medical Image Computing, Department of Computer Science, University College London, London, United Kingdom, 3Champalimaud Research, Champalimaud Centre for the Unknown, Lisbon, Portugal, 4Centre for Medical Imaging, University College London, London, United Kingdom, 5Department of Brain and Behavioural Sciences, University of Pavia, Pavia, Italy, 6Brain MRI 3T Center, IRCCS Mondino Foundation, Pavia, Italy
Synopsis
This work introduces the “Select and retrieve via direct up-sampling” network (SARDU-Net), a new method for model-free, data-driven quantitative MRI (qMRI) experiment design. SARDU-Net identifies informative measurements within lengthy acquisitions and reconstructs fully-sampled signals from a sub-protocol, without prior information on the MRI contrast. It combines two deep networks: a selector, which selects a signal sub-sample, and a predictor, which retrieves input signals. SARDU-Net can be run with standard computational resources and can increase the clinical appeal of qMRI. Here we demonstrate its potential on qMRI of prostate and spinal cord, two areas where fast acquisitions are key.
Introduction
The latest quantitative MRI (qMRI) acquisitions have increased dramatically the number of images1 and contrasts2 that can be acquired per unit time. However, the increased acquisition complexity makes the design of clinically viable protocols challenging. Clinical acquisitions need to capture salient characteristics of vast sampling spaces within acceptable times, trading off between scan duration and protocol richness3.Here we tackle the issue of designing compact samplings in vast acquisition spaces by introducing the “Select and Retrieve via Direct Up-sampling” network (SARDU-Net), which identifies informative measurements within lengthy qMRI acquisitions. Unlike state-of-the-art statistical methods as those based on Fisher information4, SARDU-Net does not rely on any explicit signal model/representation5,6. We demonstrate its potential on data from prostate and spinal cord, two areas difficult to image due to physiological noise7,8, for which rapid acquisitions are imperative.
Methods
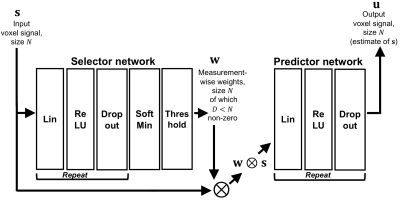
AlgorithmWe implemented SARDU-Net in PyTorch using two fully-connected deep neural networks (DNNs) (Figure 1). The first DNN is a selector, taking as input a measured voxel signal s and selecting a sub-set w⨂s, where weights w nullify non-relevant measurements. The second DNN is a predictor, outputting u, an estimate of s obtained from w⨂s. The two DNNs are optimised jointly end-to-end9 by minimising $$$L = ||\bf u - \bf s||_2^2$$$, so that the selected sub-set is the one enabling the best reconstruction of the input signals.
Acquisition
Prostate We studied scans from 7 men referred for prostate MRI following suspicion of cancer within the INNOVATE trial10. We focussed on diffusion-weighting imaging (DWI) (VERDICT11 protocol: b={0, 90, 500, 1500, 2000, 3000} s/mm2; resolution: 1.25×1.25×5 mm3) and multi-echo luminal water imaging12 (LWI) (32 echoes; TEmin/echo spacing: 31.25/31.25 ms/ms; resolution: 0.94×0.94×4.4 mm3), acquired on a 3T Philips Achieva.
Spinal cord We studied spinal cord ZOOM-EPI13 scans acquired on 4 healthy volunteers (2 males, scan-rescan) with a 3T Philips Achieva (resolution: 1×1×5 mm3), consisting of 98 measurements: 68 DWIs14 (b={0, 300, 1000, 2000, 2800} s/mm2); 10 inversion recovery images15 (inversion times in [100; 2300] ms); 11 magnetisation transfer images15 (11 off-resonance saturations); 7 multiple-TE images (mTE, TE={25, 40, 55, 70, 85, 100, 200} ms).
Post-processing
Prostate DWI was co-registered non-linearly to LWI using NiftyReg16, where a prostate mask was segmented manually. Concatenating 10 DWIs-32 LWIs provided 42 measurements.
Spinal cord We corrected data for motion and segmented the cord with SCT17.
Training
For both data sets we used SARDU-Net to select informative sub-protocols. As a test case we searched sub-protocols that are 50% (half) and 33% (a third) of the original length, although end users could search for any length.
We trained SARDU-Net for 1000 epochs using 80%-20% of prostate/cord voxels as training-validation sets (4 hidden layers for selector/predictor; learning rates: 0.001/0.0001 for 50%-33% task). We adopted a leave-one-out strategy, training in turn a SARDU-Net on all subjects but one, left out for testing.
Analysis
We tested whether SARDU-Net selects sub-protocols that capture salient characteristics of input signals. Firstly, we evaluated popular metrics from SARDU-Net signal reconstructions, namely: apparent diffusion coefficient (ADC), kurtosis (K), luminal water fraction (LWF) (prostate); fractional anisotropy (FA), T1, T2, magnetisation transfer ratio (MTR) (spinal cord). Afterwards, we compared those metrics to their counterparts obtained from the original data by bootstrapping median errors across subjects (i.e. median(mSARDU – mTRUE), with m representing a metric; 1000 bootstraps).
Results
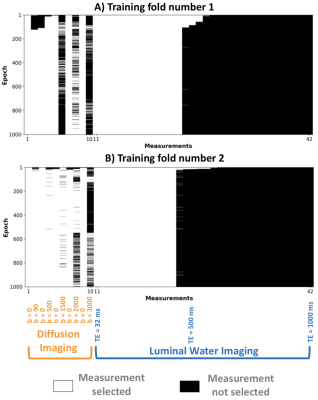
Prostate imagingFigure 2 shows sub-protocol selections in two leave-one-out folds. SARDU-Net selects low/intermediate b-values (DWI) and TE up to about 500 ms (LWI). Results are consistent across folds. Training takes ~1sec per epoch on a 8-core, 2.7 GHz Intel Core i7 processor.
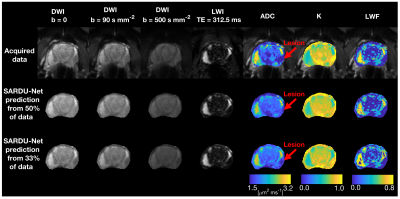
Figure 3 shows SARDU-Net image reconstructions and quantitative metrics. Reconstructed images resemble the acquired data on visual inspection and support accurate model fitting. We do not detect significant biases in reconstructed ADC, K, LWF (error confidence intervals contain error=0).
Spinal cord imaging
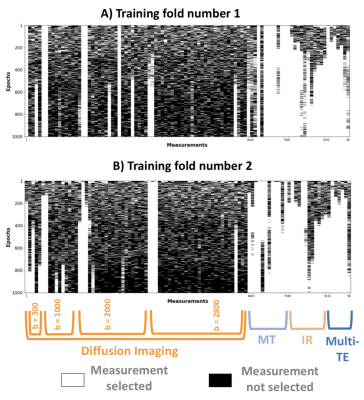
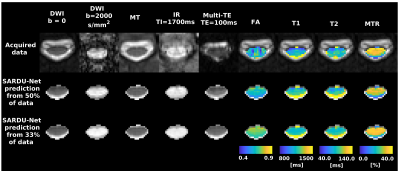
Figure 4 shows sub-protocol selections in two leave-one-out folds. SARDU-Net sub-samples heavily DWI, the longest scan, and selects measurements from all of MT, IR, mTE. Computational time per epoch is comparable to the prostate data.
Figure 5 shows SARDU-Net reconstructions and quantitative metrics. There is good correspondence between predicted and acquired data. We do not detect significant biases in FA, T1, T2, MTR (error confidence intervals contain error=0), although fine-scale anatomical details are blurred as sub-sampling gets stronger.
Discussion
In this work we introduce SARDU-Net, a deep learning tool for model-free, data-driven qMRI experiment design. SARDU-net detects informative measurements within vast acquisition spaces where brute-force searches are intractable. It does not rely on any prior MRI signal model, and can be run on commonly available resources.SARDU-Net can be trained on both simulated and in vivo data, and is ideally placed to guide the design of large clinical studies from a small number of test scans. Here we demonstrate it by selecting informative sub-protocols within lengthy prostate and spinal cord qMRI, shortening scan time with minimal information losses.
Conclusion
SARDU-Net gives new opportunity to identify economical but informative data sets for clinical applications under high time pressure. Using a small number of rich acquisitions from a pilot study with long acquisition times, it identifies the most informative sub-protocol given a specific time constraint.Acknowledgements
This project has received funding under the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 634541 and 666992, and from: Engineering and Physical Sciences Research Council (EPSRC EP/R006032/1, M020533/1, G007748, I027084, N018702); Prostate Cancer UK Targeted Call 2014: Translational Research St.2, project reference PG14-018-TR2; Spinal Research (UK), Wings for Life (Austria), Craig H. Neilsen Foundation (USA) (jointly funding the INSPIRED study); Wings for Life (#169111); UK Multiple Sclerosis Society (grants 892/08 and 77/2017); the Department of Health's National Institute for Health Research (NIHR) Biomedical Research Centres and UCLH NIHR Biomedical Research Centre; Champalimaud Centre for the Unknown, Lisbon (Portugal). We thank Philips Healthcare for assistance in protocol development and for access to research protocols.References
1. Barth M, Breuer F, Koopmans JP, et al. Simultaneous multislice (SMS) imaging techniques. Magn Reson Med 2016; 75(1):63–81.
2. Hutter J, Slator PJ, Christiaens D, et al. Integrated and efficient diffusion-relaxometry using ZEBRA. Scientific Reports 2018; 8: 15138.
3. Cercignani M, Dowell NG, Tofts PS. Quantitative MRI of the brain: principles of physical measurement. Taylor and Francis 2018, ISBN 978-1-138-03285-9.
4. Alexander DC. A general framework for experiment design in diffusion MRI and its application in measuring direct tissue-microstructure features. Magn Reson Med 2008, 60(2): 439-448.
5. Filipiak P, Fick R, Petiet A et al. Reducing the number of samples in spatiotemporal dMRI acquisition design. Magn Reson Med 2019; 81(5): 3218-3233.
6. Benjamini D, Basser PJ. Use of marginal distributions constrained optimization (MADCO) for accelerated 2D MRI relaxometry and diffusometry. Journal of Magnetic Resonance 2016, 271: 40-45.
7. Haider MA, Yao X, Finelli A et al. Multiparametric magnetic resonance imaging in the diagnosis of prostate cancer: a systematic review. Clinical Oncology 2016; 28(9): 550-567.
8. Stroman PW, Wheeler-Kingshott C, Bacon M et al. The current state-of-the-art of spinal cord imaging: Methods. NeuroImage 2014; 84: 1070-1081.
9. Bahadir CD, Dalca AV and Sabuncu MR. Learning-based Optimization of the Under-sampling Pattern in MRI. arXiv 2019: 1901.01960 [eess.IV].
10. Johnston E, Pye E, Bonet-Carne E et al. INNOVATE: A prospective cohort study combining serum and urinary biomarkers with novel diffusion-weighted magnetic resonance imaging for the prediction and characterization of prostate cancer. BMC Cancer 2016; 16(1): 816-826.
11. Panagiotaki E, Chan R, Dikaios N et al. Microstructural characterization of normal and malignant human prostate tissue with Vascular, Extracellular, and Restricted Diffusion for Cytometry in Tumours Magnetic Resonance Imaging. Investigative Radiology 2015, 50(4): 218-227.
12. Sabouri S, Chang SD, Savdie R et al. Luminal Water Imaging: A New MR Imaging T2 Mapping Technique for Prostate Cancer Diagnosis. Radiology 2017; 284(2):451-459.
13. Wheeler-Kingshott CA, Parker GJ, Symms MR et al. ADC mapping of the human optic nerve: increased resolution, coverage, and reliability with CSF-suppressed ZOOM-EPI. Magn Reson Med 2002; 47(1): 24-31.
14. Grussu F, Schneider T, Zhang H et al. Neurite orientation dispersion and density imaging of the healthy cervical spinal cord in vivo. NeuroImage 2015; 111: 590-601.
15. Battiston M, Grussu F, Ianus A et al. An optimized framework for quantitative magnetization transfer imaging of the cervical spinal cord in vivo. Magn Reson Med 2018; 79(5): 2576-2588.
16. Modat M, Ridgway GR, Taylor ZA et al. Fast free-form deformation using graphics processing units. Comput Methods Programs Biomed. 2010; 98(3): 278-84.
17. De Leener B, Lévy S, Dupont SM et al. SCT: Spinal Cord Toolbox, an open-source software for processing spinal cord MRI data. NeuroImage 2017; 145(Pt A): 24-43.
Figures