1028
Multi-path Deformable Convolutional Neural Network with Label Distribution Learning for Fetal Brain Age Prediction1Department of Radiology and BRIC, The University of North Carolina at Chapel Hill, Chapel Hill, CA, United States, 2School of Electronic and Information Engineering, South China University of Technology, GUANGZHOU, China, 3Department of Radiology, Obstetrics and Gynecology Hospital, Fudan University, ShangHai, China
Synopsis
In this study, an end-to-end framework, combining deformable convolution and label distribution learning, is developed for fetal brain age prediction based on MRI. Furthermore, a multi-path architecture is proposed to deal with multi-view MRI scenarios. Experiments on the collected dataset demonstrate that the proposed model achieves promising performance.
Introduction
Fetal magnetic resonance imaging (MRI) is being increasingly performed as part of the prenatal care to better visualize the developing brain and detect abnormalities, since it is a safe and non-invasive way to evaluate the fetal brain with greater details compared to ultrasound imaging. However, the locations and directions of the fetal brain are randomly variable and disturbed by adjacent organs, which imposes great challenges to the fetal brain age prediction. The aim of this study is to employ deformable convolution [2] and label distribution learning [3] to develop an effective framework that is capable of learning free deformations for fetal brains with different scales, shapes and directions.Methods
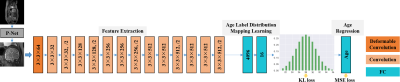
The proposed framework is based on VGG-16 [4], and we modify some architecture components to make it more suitable for fetal age prediction. This model consists of cascaded deformable and standard convolution, pooling and fully-connected layers. ReLU activation function is added after every convolution layer for nonlinear operation. Layers of this architecture are designed to implement three sequential operations for fetal brain age prediction, i.e., feature extraction, age label distribution mapping, and age regression. We implement these steps by an end-to-end network as shown in Fig. 1.To address the issue that the location of the fetal brain is randomly variable and the shape of the brain is complex, we no longer fix the regular structure of the kernel for convolution, so that the convolution can easily pay attention to interesting regions, and further capture essential structure features of the brain. Specifically, to enable adaptive localization for brains with different shapes, we apply deformable convolution, which adds offsets to the regular grid sampling location in the standard convolution, in the first convolution layer of the framework. Since direct regression of the precise age label is extremely complex, especially when the training data are not sufficient, we turn to regress the probability distribution of age first, which effectively leverages the label ambiguity in both feature learning and classifier learning, thus helping prevent the network from overfitting. The fully-connected layer has full connections to all activations in the previous layer, which can fuse feature information globally. Hence, two fully-connected layers are inserted between feature learning layers and the age regression layer, to transfer features from the fetal brain image to the age distribution. The age regression layer is responsible for transforming the distribution of age to an explicit age value. Here we utilize a fully-connected layer to learn the mapping relationship. Meanwhile, we introduce an intermediate supervision into the network to guarantee the discrepancy between the ground-truth age distribution and the predicted one. For measurement, we adopt Kullback-Leibler divergence. In addition, we control the difference between the final regressed age and the corresponding ground truth, implemented by a Mean Square Error (MSE) loss.
To make our network scalable to multi-view (sagittal, coronal, and axial views) scenarios, we train three models for each of these views and perform a comprehensive evaluation on two fusion strategies:
Late fusion: Directly averaging the predicted ages from three models as the final estimated age.
Early fusion: Multi-view images go through their own view-specific network and their estimated age probability distributions are merged with element-wise maximum.
We use de-identified fetal brain MRI stacks from 289 subjects, whose ages range from 21 weeks to 36 weeks. Each MRI stack is manually checked by an expert to make sure that the fetal brain develops normally. We use P-Net [6] to detect the center of the fetal brain and randomly crop out several expanded images containing the fetal brain from every stack. Moreover, we use Gaussian distribution to label the probability distribution of age, where the standard deviation σ is set to 0.6. This age prediction algorithms is evaluated by 3 times of 4-fold cross validation with 2 criteria, i.e., the mean absolute error (MAE) and the R2 score between the predicted ages and the ground-truth ages.
Results
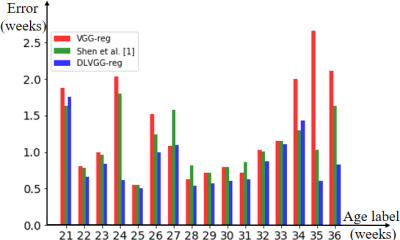
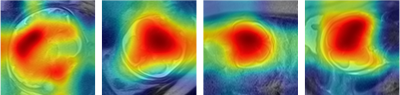
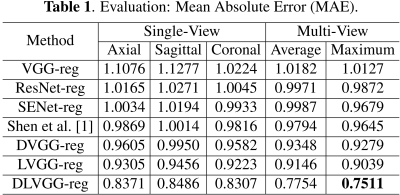
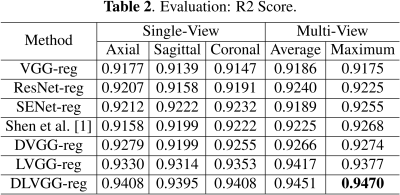
The predicted results are presented in Table 1 and Table 2. By leveraging both the deformable convolution and label distribution learning, we achieve the state-of-the-art results including a R2 score of 0.947 and mean error of 0.751 weeks. Fig. 2 shows the error of predicted age of three models for the corresponding age label. To help us better understand how the network learn effective features for fetal brain age prediction, we visualize the age activation maps of this architecture with the Grad-CAM algorithm [5]. Fig. 3 lists activation maps of four age classes.Conclusion
In this study, we present an end-to-end framework based on deformable convolution for fetal age prediction. To deal with insufficient training data, label distribution learning is introduced into our network. Furthermore, we exploit the suitable ways to address multi-view scenarios. Experiments on the collected dataset demonstrate that the proposed model significantly improves the performance on MRI based fetal age prediction.Acknowledgements
No acknowledgement found.References
1. Shen, Liyue, et al. "Deep Learning with Attention to Predict Gestational Age of the Fetal Brain." arXiv preprint arXiv:1812.07102 (2018).
2. Dai, Jifeng, et al. "Deformable convolutional networks." Proceedings of the IEEE international conference on computer vision. 2017.
3. Geng, Xin. "Label distribution learning." IEEE Transactions on Knowledge and Data Engineering 28.7 (2016): 1734-1748.
4. Simonyan, Karen, and Andrew Zisserman. "Very deep convolutional networks for large-scale image recognition." arXiv preprint arXiv:1409.1556 (2014).
5. Selvaraju, Ramprasaath R., et al. "Grad-cam: Visual explanations from deep networks via gradient-based localization." Proceedings of the IEEE International Conference on Computer Vision. 2017.
6. Ebner, Michael, et al. "An automated localization, segmentation and reconstruction framework for fetal brain MRI." International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2018.
Figures