1016
Identification and Correction of Errors in Quantitative Multi-Parameter Mapping (MPM)1Institute for Systems Neuroscience, University Medical Center Hamburg-Eppendorf, Hamburg, Germany, 2Department of Neurophysics, Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany, 3Department of Psychiatry and Psychotherapy, University Medical Center Hamburg-Eppendorf, Hamburg, Germany, 4Wellcome Trust Centre for Neuroimaging, UCL Institute of Neurology, UCL, London, United Kingdom, 5Center for Lifespan Psychology, Max Planck Institute for Human Development, Berlin, Germany, 6Laboratory for Research in Neuroimaging, Department of Clinical Neuroscience, Lausanne, Switzerland, 7Stochastic Algorithms and Nonparametric Statistics, Weierstrass Institute for Applied Analysis and Stochastics, Berlin, Germany, 8Institute of Cognitive Neurology and Dementia Research, Otto-von-Guericke-University, Magdeburg, Germany, 9German Center for Neurodegenerative Diseases, Magdeburg, Germany
Synopsis
We introduced novel error maps for proton density, longitudinal relaxation and magnetization transfer saturation rates that are more sensitive to artifacts than previously used error measures. We showed that they can be used to identify and down weigh local errors in the quantitative parameter maps for an experiment consisting of two successive multi-parameter mapping (MPM) measurements in a group of 10 healthy subjects.
Introduction
In contrast to standard weighted images, Multi-Parameter Mapping (MPM)1,2 provides multiple quantitative markers (longitudinal relaxation rate R1, proton density PD, effective transverse relaxation rate R2*, and magnetization transfer saturation MT). These parameter maps are more specific to key biological features, e.g. myelin and iron content3. However, since requiring the combination of multiple multi-echo fast-low-angle-shot (FLASH6) volumes, MPMs are also more susceptible to subject motion than traditional morphometric features such as volumetry from weighted images. In addition, the parameter estimation is based on the nonlinear relation between acquired signal and quantitative model parameters, which can amplify errors in the underlying FLASH data, e.g. caused by subject motion or physiological noise.In this study, we introduce a method to calculate error maps that are sensitive to erroneous regions in the quantiatiave MPM maps (Fig.1). For correction of errors, we introduce a method to calculate a weighted average over repeated MPMs using voxel-based weights reflecting local parameter error.
Methods
Subjects: We included 10 healthy participants in the reported analysis (Mage=31.3 years, 7 female). Exclusion criteria were psychiatric disorders, assessed via the Mini-International Neuropsychiatric Interview4, neurological diseases, head trauma or metallic implants.MRI: Scans were performed on a 3T PRISMA MRI (Siemens Healthcare, Erlangen, Germany), using the Siemens 64-channel receiver (Rx) head-coil. Whole brain MR images were acquired using the MPM2 protocol, including calibration5 and three different weightings (MT-, PD- and T1-weighting) FLASH images. The following sequence parameters were used: isotropic resolution of (1 mm)³, flip angle (FA) of 6° (for MT- and PD-weighted) and 21° (for T1-weighted), 8 echoes (2.30 to 18.40 ms, in steps of 2.30 ms), readout bandwidth of 488 Hz/pixel, and repetition time (TR) of 24.00 ms. The weighted images were acquired in two successive runs using a 3D acceleration factor of 3 and partial Fourier (6/8) (28 min.).
Step 1: Estimation of the model-fit residuals $$$\vec{\epsilon}_{inx}$$$ for each gradient-recall echo signal with PD-, T1-, and MT-weighting ($$$inx \in \{ PDw, T1w, MTw \}$$$) using the linearized exponential signal decay : $$ (1) \vec{y}_{inx}=\vec{\underline{X}\beta}_{inx}+\vec{\epsilon}_{inx}$$ with $$$\vec{y}=\ln\begin{pmatrix}S_{inx}(TE_1) \\ \vdots \\ S_{inx}(TE_N)\end{pmatrix}$$$, $$$\underline{X}$$$ being an $$$N \times 2$$$ matrix with rows $$$X_k = \left[ 1, TE_k \right], k\in 1,\dots,N$$$ where $$$N$$$ is the number of echoes and $$$\vec{\beta}_{inx}$$$ are regression coefficients. The root-mean-square of the model-fit residuals $$$\left(\epsilon_{inx} = rms\left(\vec{\epsilon}_{inx}\right)\right)$$$ was used in the next steps.
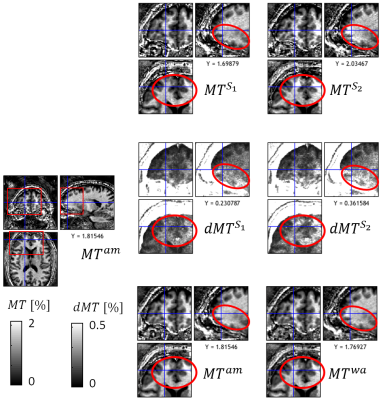
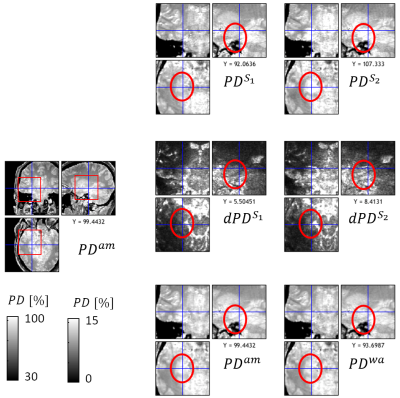
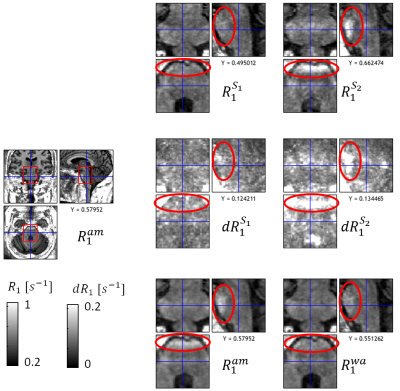
Step 2: To estimate the error of each quantitative map (Fig.1), we calculated the propagation of error for uncorrelated errors (for a function $$$F(x,y)$$$, it is defined as follows: $$$dF(x,y)=\sqrt{\left(\frac{dF}{dx}\right)^2 dx^2+\left(\frac{dF}{dy}\right)^2dy^2}$$$ ). For example, the error for the example of R1 was defined as follows: $$ (2) dR_1=\sqrt{\left(\frac{dR_1}{dS_{T1w}}\epsilon_{T1w}\right)^2+\left(\frac{dR_1}{dS_{PDw}}\epsilon_{PDw}\right)^2}$$ We used Equation (A6) in7 to calculate the derivatives of R1. The variation in the signal was approximated as follows: $$$dS_{T1w}\approx\epsilon_{T1w}$$$ and $$$dS_{PDw}\approx\epsilon_{PDw}$$$ using the corresponding residuals in Eq. (1). Estimates of ST1w and SPDw were taken as the corresponding beta-estimates at $$$TE = 0$$$.
Step 3: The two runs (S1,2) of our measurement were combined using the arithmetic mean (superscript “am”) and a weighted average (superscript “wa”). For the example of R1, the weighted-combination was defined as follows: $$(3) R_1 ^{wa} = \frac{R_1^{S1}f(ndR_1^{S1})+R_1^{S2}f(ndR_1^{S2})}{f(ndR_1^{S1})+f(ndR_1^{S2})} $$ where $$$ndR_1^{S1,2}=\frac{dR_1^{S1,2}}{dR_1^{S1}+dR_1^{S2}} $$$ and $$$f(x)=\frac{2}{exp((abs(x)-x_0)k_0)+1}$$$ being a Fermi function with two parameters ($$$x_0,k_0$$$), tuning the sensitivity of the weights with respect to the error in the data. Both parameters were heuristically optimized for one dataset that showed the worst motion artifacts.
Group analyses: The hMRI toolbox7 was used to transform MPM maps into common space. Then, the variability across subjects was assessed by generating standard-error-of-the-mean (SEM) maps of each dataset (S1, S2, arithmetic mean, AM, and weighted average, WA) and MPM parameter (Fig.5a). The relative SEM with respect to the arithmetic mean ($$$rSEM=1-SEM_{inx}/SEM_{AM}$$$ with $$$inx=\{S_1,S_2,WM\}$$$) was calculated for all datasets. Since the bias introduced by subject motion and physiology increases the variability of the estimated parameters, positive rSEM values were interpreted as improved sensitivity for group differences.
Results
The proposed error maps (middle column, Fig.1) were more sensitive and specific to the artifacts in the different quantitative MPM maps (right column) than the regular model fit residuals from Eq. (1) (left column). Moreover, the error maps clearly differed between the different parameters (rows in Fig.1). We found for all three contrasts that the arithmetic mean of the proposed protocol showed less artifacts than each acquisition separately, both locally (Fig.2-4 and blue regions in Fig.5b with up to 50% improvement) and across the whole white matter (Fig.5c, up to 20%). In addition, the weighted average showed better performance than the arithmetic mean, both at a local (red regions in Fig.5b, up to 20%) and global level (Fig.5c, 1-3%).Discussion and Conclusion
We introduced parameter specific error maps for the quantitative MPM maps (R1, PD, MT) that are more specific and sensitive than previously introduced error maps (Eq. (1) and8). The error maps can be used to down-weigh erroneous regions. We found that their arithmetic average clearly improved the stability of the quantitative MPM maps, and that the proposed weighted-average further reduced variation. Future investigation will explore how the results generalize when larger cohorts are investigated and the suitability for patient groups.Acknowledgements
This project was supported by LPEK_006_Kühn, by the ERA-NET NEURON (hMRI- ofSCI), the Bundesministerium für Bildung und Forschung (BMBF; 01EW1711A and B), and the Deutsche Forschungsgemeinschaft (grant MO 2397/4-1), and the Forschungszentrums Medizintechnik Hamburg (fmthh; grant 01fmthh2017).References
1. Helms, G., Dathe, H. & Dechent, P. Quantitative FLASH MRI at 3T using a rational approximation of the Ernst equation. Magn Reson Med 59, 667–672 (2008).
2. Weiskopf, N. et al. Quantitative multi-parameter mapping of R1, PD*, MT and R2* at 3T: a multi-center validation. Front. Neurosci. 7:, 95 (2013).
3. Callaghan, M. F., Helms, G., Lutti, A., Mohammadi, S. & Weiskopf, N. A general linear relaxometry model of R1 using imaging data. Magn Reson Med 73, 1309–1314 (2015).
4. Ackenheil, M., Stotz-Ingenlath, G., Dietz-Bauer, R. & Vossen, A. MINI Mini International Neuropsychiatric Interview, German Version 5.0.0, DSM IV. Psychiatrische Universit{ä}tsklinik M{ü}nchen, Germany (1999) doi:10.1016/0968-0004(94)90058-2.
5. Lutti, A. et al. Robust and fast whole brain mapping of the RF transmit field B1 at 7T. PLoS ONE 7, e32379 (2012).
6. Frahm, J., Haase, A. & Matthaei, D. Rapid three-dimensional MR imaging using the FLASH technique. J Comput Assist Tomogr 10, 363–368 (1986).
7. Tabelow, K. et al. hMRI – A toolbox for quantitative MRI in neuroscience and clinical research. NeuroImage 194, 191–210 (2019).
8. Weiskopf, N., Callaghan, M. F., Josephs, O., Lutti, A. & Mohammadi, S. Estimating the apparent transverse relaxation time (R2*) from images with different contrasts (ESTATICS) reduces motion artifacts. Front. Neurosci 8, 278 (2014).
Figures