1007
Reproducibility and Repeatability of Three-dimensional Magnetic Resonance Fingerprinting-based Human Brain Morphometry1Department of Radiology, Juntendo University, Tokyo, Japan, 2Department of Radiology, The University of Tokyo, Tokyo, Japan, 3Imago7 Foundation, Pisa, Italy, 4IRCCS Stella Maris, Paris, Italy, 5Department of Physics, University of Pisa, Pisa, Italy, 6MR Applications and Workflow, GE Healthcare, Tokyo, Japan, 7GE Healthcare, Munich, Germany, 8Department of Radiological Sciences, Tokyo Metropolitan University, Tokyo, Japan, 9Department of Radiology, Toho University Omori Medical Center, Tokyo, Japan
Synopsis
Magnetic Resonance fingerprinting (MRF) provides simultaneous acquisition of T1 and T2 values with high reliability. However, the reproducibility and repeatability of human brain morphometry based on MRF still requires investigation. Here, we examined the feasibility of three-dimensional (3-D) MRF to evaluate the brain cortical thickness and volumetric analysis in healthy volunteers. Scan-rescan tests of both 3-D MRF and conventional 3D T1-weighted imaging were performed. For each sequence, the regional cortical thickness and volume of the subcortical structures were measured using automatic brain segmentation software. High agreement between conventional scans and scan-rescan repeatability in healthy human brains were observed.
Introduction
Changes in the brain morphometry, such as regional cortical thickness and subcortical volumes, are reportedly associated with development, aging, and a wide variety of psychiatric and neurological disorders.1 These morphological measures can quantify the neuroanatomical features, and these accurate measurements could enable better understanding of individual characteristics, including clinical, behavioral, and genetic measures.2Magnetic resonance fingerprinting (MRF) is a multiparametric imaging technique that provides simultaneous acquisition of MR properties with single acquisition.3 Previous in vitro and in vivo studies have shown the high reproducibility and repeatability of T1 and T2 values obtained with MRF.4,5 However, studies focusing on reliability of the brain morphometry derived from MRF have been limited despite its clinical importance. Therefore, the purpose of this study was (1) to evaluate the reproducibility of three-dimensional (3-D) MRF-derived morphological measurements using conventional T1-weighted structural imaging-derived measurements as reference standards, and (2) to evaluate the repeatability of 3-D MRF-derived measurements with scan-rescan tests.
Methods
This study included 12 healthy volunteers (4 females and 8 males; age range 22-61 years). All participants were scanned using a 3-T scanner (Discovery 750 w, GE Healthcare, Waukesha, WI, USA) with a 32-channel head coil. Scan-rescan tests of both 3-D MRF and 3-D T1-weighted fast spoiled gradient recalled echo (FSPGR) sequence were performed for all the participants. Between the scan and rescan tests, the subjects took a short break outside the MRI room. The 3-D MRF sequence was based on steady-state free precession (SSFP) with spiral projection k-space.6 The acquisition parameters of the MRF were as follows: field-of-view (FOV) 200×200×200 mm; matrix size 200×200×200 mm; acquisition time was 9 min 51 sec. The dictionary was generated using the extended phase graphs formalism, with T1 value ranging from 10ms to 100ms with 10ms steps, 100ms to 1s with 100ms steps, 1s to 2s with 50ms steps, and 2s to 6s with 100ms steps. MRF T1 maps were obtained by inner-product pattern matching. MRF-derived T1 maps were post-processed voxel-wisely to generate synthetic T1-weighted images by the following formula:$$S=1-2e^{-TI/T1}$$
where, S is the output signal, T1 is T1 value obtained by MRF, and TI is the inversion time virtually set to 1300ms. The imaging parameters of FSPGR were sagittal acquisition; TR/ TE/ TI, 7.7/3.1/400ms; FOV 256×256 mm; matrix size 256×256 mm; section thickness 1.0mm; flip angle 11°; receiver bandwidth 244.1 Hz/pixel; and acquisition time 5 min 45 sec. One participant was excluded due to motion artifacts exhibited in both 3-D MRF and FSPGR. Each anatomical volume was analyzed using the FreeSurfer (Version 6.0) 7 to obtain individual-specific regional cortical thickness in the Desikan-Killiany Atlas.8 The FMRIB Integrated Registration and Segmentation Tool (FIRST) was used to obtain volumes of the subcortical structures.9 Percent relative difference and intraclass correlation coefficient (ICC) were used to assess the reproducibility between the 3-D MRF and FSPGR sequence-derived measurements. Within-subject coefficient of variation (wCV) was also used in assessing the repeatability for both sequences.
Results
Representative FreeSurfer and FIRST outputs from 3-D MRF sequence-derived T1-weighted images are shown in Figure 1.Measurement of Cortical Thickness
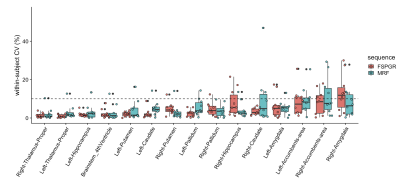
The percent relative difference in cortical thickness across all regions with FSPGR was 9.6 ± 3.7% (Figure 2). The ICC across all regions was 0.68 ± 0.25, and 59% of the regional areas showed substantial (ICC > 0.61) to almost perfect agreement (ICC > 0.81). The wCV of cortical thickness across all regions with 3-D MRF was 1.5 ± 0.9%, while that with FSPGR was 1.3 ± 1.0% (Figure 3).
Volumetry of Subcortical Structures
Percent relative difference in the volume of subcortical structures across all structures with FSPGR was 8.1 ± 4.6% (Figure 4). The ICC across all regions was 0.80 ± 0.15, and all regions except the amygdala showed substantial to almost perfect agreement. The wCV of the volume of subcortical structures across all structures with 3-D MRF was 4.0 ± 2.5%, while that with FSPGR was 4.2 ± 3.5% (Figure 5).
Discussion
3-D MRF-derived regional cortical thickness and subcortical volumes showed high repeatability and good agreement with the FSPGR-derived measurements. To our knowledge, this is the first study to evaluate the reliability of 3-D MRF-derived morphometry of the human brain.Accurate volumetric segmentation of the brain is a prerequisite to detect subtle regional changes in quantitative T1 and T2 values. Our results demonstrated that 3-D MRF provides anatomical information that is as reliable as conventional 3-D T1-weighted imaging. Along with the highly reliable T1 and T2 value quantification, 3-D MRF can identify subtle regional changes that may have been obscured while observing a large region.
We used T1-weighted images with fixed TI for segmentation in this study. Optimizing parameters could further improve the segmentation performance. Additionally, using multichannel inputs (e.g., T1 and T2 maps), which could be obtained from a single 3-D MRF sequence scan, have the potential to improve the robustness of segmentation algorithms that rely only on T1-weighted images.
Conclusion
Thus, 3-D MRF is as reliable as conventional 3-D T1-weighted images in measuring the cortical thickness and subcortical volumes in a healthy human brain. Further longitudinal and multicenter studies incorporating scanners from different sites and field strengths should be conducted.Acknowledgements
No acknowledgement found.References
- Fotenos AF, Snyder AZ, Girton LE, Morris JC, Buckner RL. Normative estimates of cross-sectional and longitudinal brain volume decline in aging and AD. Neurology 2005;64:1032–1039.
- Sabuncu MR, Ge T, Holmes AJ, et al. Morphometricity as a measure of the neuroanatomical signature of a trait. Proc Natl Acad Sci U S A 2016;113(39):E5749-5756.
- Ma D, Gulani V, Seiberlich N, et al. Magnetic resonance fingerprinting. Nature. 2013;495(7440):187-192.
- Buonincontri G, Biagi L, Retico A, et al. Multi-site repeatability and reproducibility of MR fingerprinting of the healthy brain at 1.5 and 3.0T. Neuroimage 2019;195:362-372.
- Korzdorfer G, Kirsch R, Liu K, et al. Reproducibility and Repeatability of MR Fingerprinting Relaxometry in the Human Brain. Radiology 2019;292(2):429-437.
- Cao X, Ye H, Liao C, Li Q, He H, Zhong J. Fast 3D brain MR fingerprinting based on multi-axis spiral projection trajectory. Magn Reson Med 2019;82(1):289-301.
- Fischl B. FreeSurfer. Neuroimage 2012;62:774–781
- Desikan RS, Segonne F, Fischl B, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 2006;31:968–980.
- Patenaude B, Smith SM, Kennedy DN, Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage 2011;56:907–922.
Figures
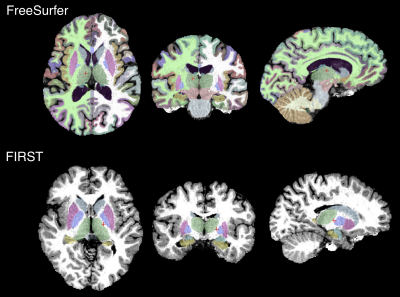
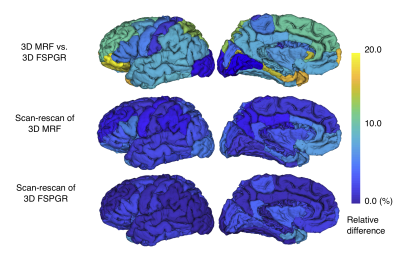
Figure 2. Percent relative difference of cortical thickness among scans
(a) Comparison between 3-D magnetic resonance fingerprinting (MRF) and fast spoiled gradient recalled echo (FSPGR); (b) and (c) Scan-rescan of 3-D MRF and FSPGR, respectively. Regional percent relative difference is overlaid on an inflated brain surface.
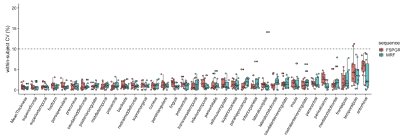
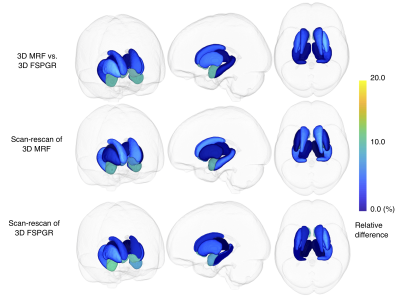
Figure 4. Percent relative difference of subcortical volumes among scans
(a) Comparison between 3-D magnetic resonance fingerprinting (MRF) and fast spoiled gradient recalled echo (FSPGR); (b) and (c) Scan-rescan of 3-D MRF and FSPGR, respectively. Each subcortical structure is colored with the regional percent relative difference, and the brain surface is made transparent for ease of visualization.