1004
FITs-CNN: A Very Deep Cascaded Convolutional Neural Networks Using Folded Image Training Strategy for Abdominal MRI Reconstruction
Satoshi Funayama1, Tetsuya Wakayama2, Hiroshi Onishi1, and Utaroh Motosugi1
1Department of Radiology, University of Yamanashi, Yamanashi, Japan, 2GE Healthcare Japan, Tokyo, Japan
1Department of Radiology, University of Yamanashi, Yamanashi, Japan, 2GE Healthcare Japan, Tokyo, Japan
Synopsis
For faster abdominal MR imaging, deep learning-based reconstruction is expected to be a powerful reconstruction method. One of the challenges in deep learning-based reconstruction is its memory consumption when it is combined with parallel imaging. To handle the problem, we propose a very deep cascaded convolutional neural networks (CNNs) using folded image training strategy (FITs). We also present that the network can be trained with FITs and shows good quality of reconstructed images.
Introduction
Deep learning-based reconstruction methods utilize convolutional networks (CNNs) to learn priors to enable image reconstruction from the data undersampled with high acceleration factors1,2. These methods showed the better performance and the faster abdominal MR imaging in case of a single coil image. A combination with parallel imaging may improve performance of network by utilizing the spatial sensitivity information of a phase-array coil3,4. One of the challenges in CNNs combined with parallel imaging is memory consumption, which restricts the depth of CNNs and the number of coil channels, especially in network training. To handle this, we propose a very deep cascade CNNs using folded image training strategy (FITs-CNN) to combine it with data driven parallel imaging for abdominal MRI reconstruction.Methods
Image Reconstruction: The proposed FITs-CNN pipeline is the combination of CNNs and the following data driven parallel imaging to reconstruct undersampled images. In this study, the undersampled data was prepared by the following two steps. 1) The fully sampled k-space data was uniformly undersampled with auto-calibration signal (ACS) points, which is normally used in parallel imaging. 2) For further undersampling, the remaining k-space points are pseudo-randomly undersampled with variable density Poisson disk sampling5. The learning of CNNs is performed on the folded image domain, which learns the conversion from the highly undersampled images at step 2) to uniformly undersampled image at step 1). The estimated folded images by CNNs are transformed back to k-space to fill the missed k-space data points using data driven parallel imaging (ARC)6 to obtain the final unfolded image.Network Architecture: The proposed very deep cascaded CNNs iterates between 2D CNN blocks and data consistency layers to ensure the fidelity of sampled k-space point through the iterations (Figure 1). To handle the memory consideration, the network takes the ky-kz plane images folded in ky direction, which comes from the uniform undersampling. the weights were shared among all the CNN blocks. The network is trained end-to-end based on the mean squared error between output and ground truth folded images using Adam optimizer on a NVIDIA GTX 1080 Ti.
Training Data: Fully sampled fast spoiled gradient echo images were acquired from 10 patients on abdominal MRI examinations using a 3.0-T GE scanner and 32-channel torso coil. Because the acquired complex images (256 x 256) were coil-combined, we performed an 8-channel coil simulation for multichannel reconstruction. Five volume data of ten cases were split slice-by-slice along kx (readout) direction for training, which are further augmented by random flipping and rotation. Finally, 81,920 images were used for training.
Evaluation: The other five cases not used in the training were used for evaluation. Each volume data was undersampled with total reduction factors of 2, 4, 6, 8, and 10 (random undersampling factor: 1, 2, 3, 4, 5; uniform undersampling factor: 2 for all). Image quality of reconstructed images were assessed with peak-to-noise ratio (PSNR) and structural similarity index (SSIM). Images reconstructed using zero-filling and DCCNN were used for comparison7. Un-paired t-test was used to compare the metrics differences of each method on same total reduction factor.
Results
The proposed network showed the best quality in each reduction factor compared with zero-filling and DCCNN (Figure 2). The PSNR and SSIM were significantly higher in the proposed network in total reduction factor of 8; PSNR, 44.1±1.3 vs. 42.1±1.5 (mean±SD, proposed vs. DCCNN, p<0.0001); SSIM, 0.986±0.004 vs. 0.980±0.005 (proposed vs. DCCNN, p<0.0001) (Figure 3).Discussion
We combined the very deep cascade CNNs trained by folded images and data driven parallel imaging for abdominal MRI reconstruction. To our knowledge, it is unique to use folded images for training of network in MRI reconstruction, which enable to reduce memory consumption and yield deeper network. The memory consumption of proposed network (20 iteration blocks) is a little less than a half of DCCNN (10 iteration blocks). Our method showed significantly better performance than DCCNN. However, we need to ensure that the use of folded images for training would not have a risk of degradation of network performance in a future work. We speculate that the localized sensitivity of each coil relieves overlaps of folded images and make network performance better, but further study is needed for confirmation.Conclusion
A very deep cascaded convolutional neural networks using folded image training strategy combined with data driven parallel imaging can result in better image quality for Abdominal MRI.Acknowledgements
No acknowledgement found.References
- Akçakaya M, Moeller S, Weingärtner S, et al. Scan-specific robust artificial-neural-networks for k-space interpolation (RAKI) reconstruction: Database-free deep learning for fast imaging. Magn Reson Med. 2019;81(1):439-453.2.
- Schlemper J, Caballero J, Hajnal J V.,et al. A Deep Cascade of Convolutional Neural Networks for Dynamic MR Image Reconstruction. IEEE Trans Med Imaging. 2018;37(2):491-503.3.
- Aggarwal HK, Mani MP, Jacob M. MoDL: Model-Based Deep Learning Architecture for Inverse Problems. IEEE Trans Med Imaging. 2019;38(2):394-405.4.
- Hammernik K, Klatzer T, Kobler E, et al. Learning a variational network for reconstruction of accelerated MRI data. Magn Reson Med. 2018;79(6):3055-3071. 5.
- Mitchell SA, Rand A, Ebeida MS, et al. Variable radii poisson-disk sampling [Internet]. Proceedings of the 24th Canadian Conference on Computational Geometry, CCCG 2012.6.
- Brau ACS, Beatty PJ, Skare S, et al. Comparison of Reconstruction Accuracy and Efficiency Among Autocalibrating Data-Driven Parallel Imaging Methods. Magn Reson Med. 2006;59(2):382-395.7.
- Schlemper J, Caballero J, Hajnal J V, et al. A Deep Cascade of Convolutional Neural Networks for MR Image Reconstruction. https://arxiv.org/pdf/1703.00555.pdf.
Figures
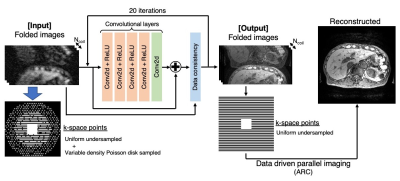
The proposed network consists of CNNs and data consistency layer. The residual CNN blocks have four CNN activated with ReLU followed by a simple 2D CNN. The data consistency layers have a function to preserve sampled k-space points. This network takes folded images, which reduce memory consumption and enable to make it deeper. Data driven parallel imaging (ARC) is applied to unwrap the folded images.
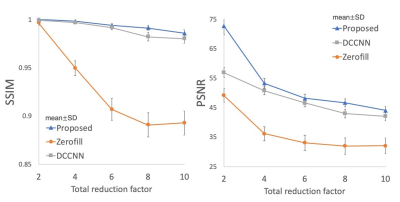
The comparison of network evaluation. The proposed network showed the better image quality in all the cases with different total reduction factor.
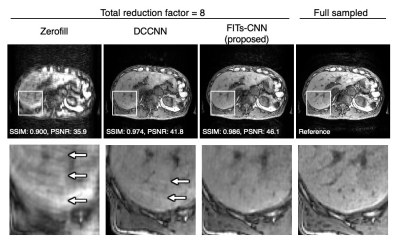
The representative liver images in the various reconstruction methods obtained from the same raw data with the total reduction factor of 8. A few aliasing artifacts was observed in DCCNN but it was removed in FITs-CNN.